Hello,
I’ve been working on modifying a version of MACE as implemented in this example Automatic rendering of Pyro models — Pyro Tutorials 1.9.1 documentation.
I added masking to it, which worked fine, but now I’m trying to modify the model to handle multiple truth values per item. The idea being that there are different categories of annotators, and for each item for each category there should be a different truth value.
In practice what I did was add a plate, so instead of my truth values, T, looking like this: tensor([[1], [2]]), now they look like this tensor([[1, 0], [2, 1]]).
For the purposes of comparing to annotations, I am now indexing T because I want to select the truth value that is associated with the annotator’s category. So if annotator 0, has category 1, then it should select the truth values at index 1 of each item. (in this example that’s 0 for the first item and 1 for the second item).
To do this indexing, I am writing T[...,categories]. Printing this out shows that it is doing the operation that I want.
Because T is a discrete variable, my understanding is that I need to block it from the autoguide, and then use enumeration; however, with the enumeration, I start getting index out of bounds errors that I didn’t get before:
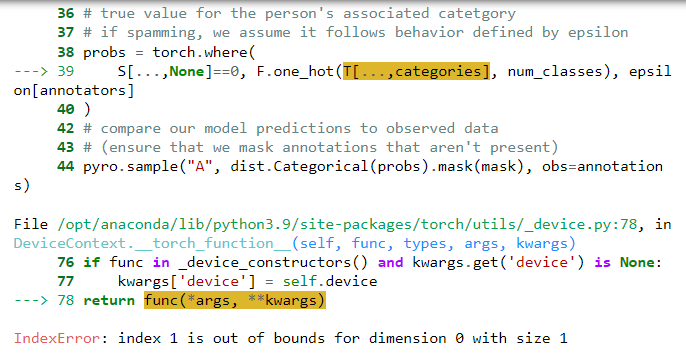
I’ve also noticed that when I have enumeration and have the model print T, the shape changes drastically. The first time it prints T looks like this tensor([[1, 0], [2, 1]]) as I expect, but the second time it looks like this tensor([[[0]], [[1]], [[2]]])
I also tried doing indexing using the pyro.ops.indexing Index and Vindex, but I have the same issues.
I have also been trying to review the documentation on enumeration, but I’m definitely missing something.
I’m certain that I’m misunderstanding something about how enumeration works. Is what I’m trying to do possible? Am I going about it the wrong way?
Here’s my code:
@config_enumerate
def model(annotators, annotations, mask, categories, alpha=0.5, beta=0.5):
# calculate shapes of our input data
num_annotators = int(torch.max(annotators)) + 1
num_classes = int(torch.max(annotations)) + 1
num_items, num_annotators = annotations.shape
num_categories = int(torch.max(categories)) + 1
# each annotator has parameters for trustworthiness and spamming behavior
with pyro.plate("annotator", num_annotators):
# theta_j is 1 - probability that annotator j is spamming
theta = pyro.sample("θ", dist.Beta(alpha, beta))
# epsilon_j is distribution of spamming behavior for annotator j
epsilon = pyro.sample("ε", dist.Dirichlet(torch.full((num_classes,), 10.)))
# for each item
with pyro.plate("item", num_items, dim=-2):
# sample different truth value for each category of annotator
with pyro.plate("category", num_categories):
T = pyro.sample("T", dist.Categorical(logits=torch.zeros(num_classes)))
# for each annotator's position on an item
with pyro.plate("position", num_annotators):
# sample whether or not they are spamming
S = pyro.sample("S", dist.Bernoulli((1 - theta[annotators])))
# if not spamming, we assume probability of the selected value is the
# true value for the person's associated catetgory
# if spamming, we assume it follows behavior defined by epsilon
probs = torch.where(
S[...,None]==0, F.one_hot(T[...,categories], num_classes), epsilon[annotators]
)
# compare our model predictions to observed data
# (ensure that we mask annotations that aren't present)
pyro.sample("A", dist.Categorical(probs).mask(mask), obs=annotations)
return T, S
# define input data
annotators = torch.tensor([0, 1, 2])
annotations = torch.tensor([[0, 1, 0],[2, 0, 2]])
mask = torch.tensor([[True, True, False], [True, True, True]])
categories = torch.tensor([0, 1, 0])
# initialize parameters
alpha, beta = 0.5, 0.5
# declare an autoguide
guide = AutoDiagonalNormal(poutine.block(model, hide=["T","S"]))
# SVI setup
optimizer = Adam({"lr": 0.1})
elbo = TraceEnum_ELBO(max_plate_nesting=2)
svi = SVI(model, guide, optimizer, loss=elbo)
# training
num_steps=1000
for step in range(num_steps):
loss = svi.step(annotators, annotations, mask, categories, alpha, beta)
if step % 100 == 0:
print(f"Step {step}, Loss = {loss}")