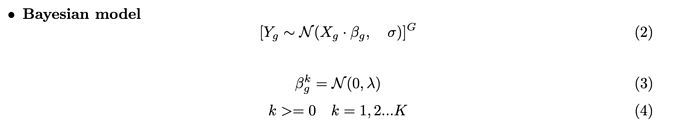I’m inferring parameters for ~20k genes independently.
this is computationally intensive, So I created a vectorised model in an attempt to avoid using a for loop to infer posterior and predict samples from held out data. but i get the error about broadcasting when predicting on held out data. maybe my model definition is flawed.
$ vectorized_samples_predictive_test = predictive(random.PRNGKey(0), geneID_CodeT, X_treatmentT, X_methylationT, None)
File "/home/ahunos/miniforge3/envs/numpyro/lib/python3.12/site-packages/numpyro/primitives.py", line 47, in apply_stack
handler.process_message(msg)
File "/home/ahunos/miniforge3/envs/numpyro/lib/python3.12/site-packages/numpyro/primitives.py", line 546, in process_message
broadcast_shape = lax.broadcast_shapes(
^^^^^^^^^^^^^^^^^^^^^
ValueError: Incompatible shapes for broadcasting: shapes=[(6,), (12,)]
--------------------
For simplicity, JAX has removed its internal frames from the traceback of the following exception. Set JAX_TRACEBACK_FILTERING=off to include these.
i’m attaching the for-loop version which works as desired and an attempt on vectorized approach. I would appreciate your help. thanks in advance
import argparse
import numpy as np
import pandas as pd
import jax.numpy as jnp
import numpyro
from numpyro.infer import MCMC, NUTS, Predictive
import numpyro.distributions as dist
from jax import random
import jax
#####for loop version of model
#y ~ B_0 + B_t * treatment + B_M*methylation
def fit_per_gene(X_treatment, X_Methylation, Y_RNA):
λ = numpyro.sample("λ", dist.LogNormal(0.0, 1.0))
σ = numpyro.sample("σ", dist.LogNormal(0.0, 1.0))
βg_t = numpyro.sample("βg_t", dist.Normal(0.0, λ))
βg_μ = numpyro.sample("βg_μ", dist.Normal(0.0, λ))
βg_M = numpyro.sample("βg_M", dist.Normal(0.0, λ))
mean_est = βg_μ + βg_t * X_treatment + βg_M * X_Methylation
with numpyro.plate("data", X_Methylation.shape[0]):
return numpyro.sample("Y_g", dist.Normal(mean_est, σ), obs=Y_RNA)
#define inference algorithm
num_warmup, num_samples, nChains= 1000, 2000, 1
def mcmc_infer_forLoop(modelName, X1, X2,Y):
# function to infer
print("running NUTS mcmc")
nuts_kernel = NUTS(model=modelName)
mcmc = MCMC(nuts_kernel, num_samples=num_samples, num_warmup=num_warmup, num_chains=nChains)
rng_key = random.PRNGKey(0)
mcmc.run(rng_key, X1, X2, Y)
posterior_samples = mcmc.get_samples()
return mcmc, posterior_samples
#pls see data
test_filtered_2 = pd.DataFrame({'geneID': {78497: 'ENSMUSG00000000001.4', 114173: 'ENSMUSG00000000001.4', 182543: 'ENSMUSG00000000028.15', 87450: 'ENSMUSG00000000028.15', 180714: 'ENSMUSG00000000056.7', 97480: 'ENSMUSG00000000056.7'}, 'geneID_Code': {78497: 0, 114173: 0, 182543: 1, 87450: 1, 180714: 2, 97480: 2}, 'treatment_code': {78497: 0, 114173: 1, 182543: 0, 87450: 1, 180714: 0, 97480: 1}, 'subjectCode': {78497: 0, 114173: 3, 182543: 1, 87450: 5, 180714: 1, 97480: 4}, 'fraction_modified_entropy_in_log2': {78497: 2.75907362977019, 114173: 3.18031794534646, 182543: 2.2957711627112, 87450: 2.92107546221738, 180714: 2.4005147597498, 97480: 2.48258530293577}, 'log_RPKM': {78497: 5.07697372927925, 114173: 4.90477412140957, 182543: 4.5252617067512, 87450: 4.23892601125065, 180714: 2.16339766406175, 97480: 2.5117697228789}})
train_filtered_2 = pd.DataFrame({'geneID': {185477: 'ENSMUSG00000000001.4', 90414: 'ENSMUSG00000000001.4', 54672: 'ENSMUSG00000000001.4', 102304: 'ENSMUSG00000000001.4', 75534: 'ENSMUSG00000000028.15', 111208: 'ENSMUSG00000000028.15', 51708: 'ENSMUSG00000000028.15', 99337: 'ENSMUSG00000000028.15', 49849: 'ENSMUSG00000000056.7', 109349: 'ENSMUSG00000000056.7', 73675: 'ENSMUSG00000000056.7', 85591: 'ENSMUSG00000000056.7'}, 'geneID_Code': {185477: 0, 90414: 0, 54672: 0, 102304: 0, 75534: 1, 111208: 1, 51708: 1, 99337: 1, 49849: 2, 109349: 2, 73675: 2, 85591: 2}, 'treatment_code': {185477: 0, 90414: 1, 54672: 0, 102304: 1, 75534: 0, 111208: 1, 51708: 0, 99337: 1, 49849: 0, 109349: 1, 73675: 0, 85591: 1}, 'subjectCode': {185477: 1, 90414: 5, 54672: 2, 102304: 4, 75534: 0, 111208: 3, 51708: 2, 99337: 4, 49849: 2, 109349: 3, 73675: 0, 85591: 5}, 'fraction_modified_entropy_in_log2': {185477: 2.25689485690177, 90414: 2.96597692903088, 54672: 3.94189649011665, 102304: 2.34897393243657, 75534: 3.3429850171158, 111208: 3.71140159408762, 51708: 3.3235481162084, 99337: 3.15423353444651, 49849: 2.98017501891044, 109349: 2.5248697814855, 73675: 2.86908906351306, 85591: 2.9733580610711}, 'log_RPKM': {185477: 5.1366364181323, 90414: 4.82371725578574, 54672: 5.05537877635977, 102304: 4.91679774620665, 75534: 4.64845582495391, 111208: 4.24286557974403, 51708: 4.63688282250396, 99337: 4.12775623116681, 49849: 2.1278698665813, 109349: 2.50706481669696, 73675: 2.25131000608453, 85591: 2.45633482627132}})
#make train arrays
geneID_Code = jnp.array(train_filtered_2['geneID_Code'].values)
X_treatment = jnp.array(train_filtered_2['treatment_code'].values)
X_methylation = jnp.array(train_filtered_2['fraction_modified_entropy_in_log2'].values)
Y_rna = jnp.array(train_filtered_2['log_RPKM'].values)
inference_holder = {}
for i, v in enumerate(np.unique(train_filtered_2["geneID_Code"].values)):
trainInfernceGeneI = {"model" : None, "mcmc" : None, "posterior_samples_Normal" : None}
print(f"model for iter={i}; geneID={v}")
data = train_filtered_2[train_filtered_2["geneID_Code"] == v]
# jnp.array(data["treatment_code"].values)
datajnp = jnp.array(data[["treatment_code", "fraction_modified_entropy_in_log2", "log_RPKM"]])
mcmc, posterior_samples_Normal = mcmc_infer_forLoop(fit_per_gene, datajnp[:, 0], datajnp[:, 1], datajnp[:, -1])
inference_holder[v] = {"model" : fit_per_gene, "mcmc" : mcmc, "posterior_samples_Normal" : posterior_samples_Normal}
testdf_gene1 = jnp.array(train_filtered_2[train_filtered_2["geneID_Code"] == 1][["treatment_code", "fraction_modified_entropy_in_log2", "log_RPKM"]])
posterior_testData_holder = {}
for i,v in enumerate(np.unique(test_filtered_2["geneID_Code"].values)):
testdata = jnp.array(train_filtered_2[train_filtered_2["geneID_Code"] == v][["treatment_code", "fraction_modified_entropy_in_log2", "log_RPKM"]])
predictive_test = Predictive(model = fit_per_gene, posterior_samples=inference_holder[v]['posterior_samples_Normal'])
samples_predictive_test = predictive_test(random.PRNGKey(0), testdata[:, 0], testdata[:, 1], Y_RNA=None)
posterior_testData_holder[v] = {"samples_predictive_test" : samples_predictive_test}
################## vectorized version of model ##################
def vectorizedNormal_model(geneID_Code, X_treatment, X_methylation, Y_RNA=None):
n_genes = len(np.unique(geneID_Code))
λ = numpyro.sample("λ", dist.LogNormal(0.0, 1.0))
σ = numpyro.sample("σ", dist.LogNormal(0.0, 1.0))
with numpyro.plate(str("g = 1,2..G"), n_genes):
βg_M = numpyro.sample("βg_M", dist.Normal(0.0, λ))
βg_μ = numpyro.sample("βg_μ", dist.Normal(0.0, λ))
βg_t = numpyro.sample("βg_t", dist.Normal(0.0, λ))
RNA_μ̂ = numpyro.deterministic("RNA_μ̂", βg_μ[geneID_Code] + βg_t[geneID_Code] * X_treatment + βg_M[geneID_Code] * X_methylation)
with numpyro.plate("obs_RNA", X_treatment.shape[0]):
obs = numpyro.sample("Yg", dist.Normal(RNA_μ̂, σ), obs=Y_RNA)
#inference
nuts_kernel = NUTS(model=vectorizedNormal_model)
mcmc = MCMC(nuts_kernel, num_samples=100, num_warmup=100, num_chains=1)
rng_key = random.PRNGKey(2)
mcmc.run(rng_key, geneID_Code, X_treatment, X_methylation, Y_rna)
posterior_samples = mcmc.get_samples()
# posterior_samples.keys()
# test_filtered_2 = test_filtered[test_filtered["geneID_Code"].isin([0, 1, 2])]
geneID_CodeT = jnp.array(test_filtered_2['geneID_Code'].values)
X_treatmentT = jnp.array(test_filtered_2['treatment_code'].values)
X_methylationT = jnp.array(test_filtered_2['fraction_modified_entropy_in_log2'].values)
Y_rnaT = jnp.array(test_filtered_2['log_RPKM'].values)
predictive = Predictive(model = vectorizedNormal_model, posterior_samples=posterior_samples)
vectorized_samples_predictive_train = predictive(random.PRNGKey(0), geneID_Code, X_treatment, X_methylation, None)
vectorized_samples_predictive_test = predictive(random.PRNGKey(0), geneID_CodeT, X_treatmentT, X_methylationT, None) #error here