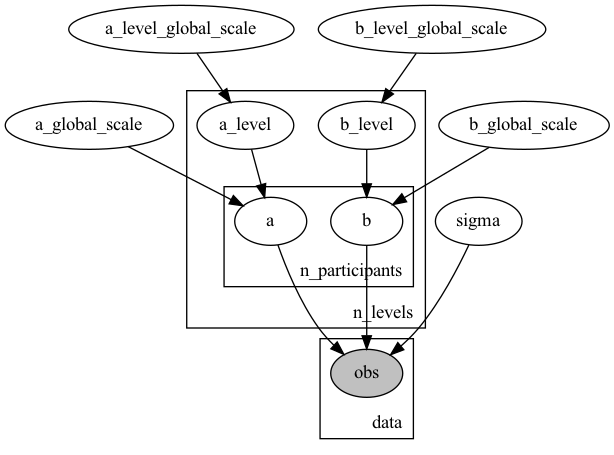
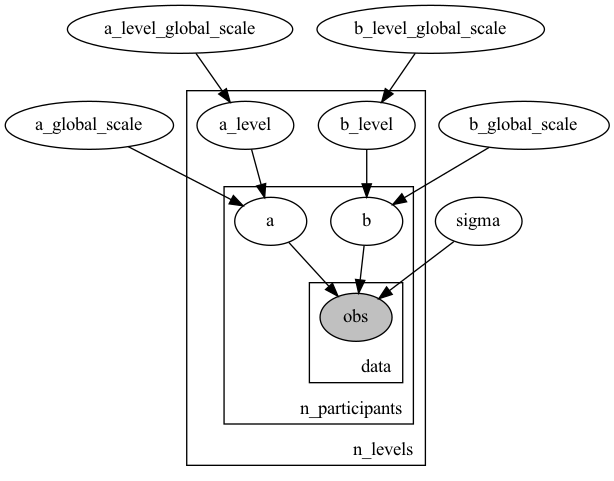
How do I use nested plate notation to build a hierarchical model for dataset that looks like this:
below : intensity_i_jk, output_i_jk represent the i’th observation for (participant_j, level_k) combination
participant, level, intensity, output
participant_1, level_1, intensity_1_11, output_1_11
participant_1, level_1, intensity_2_11, output_2_11,
...
participant_1, level_1, intensity_100_11, output_100_11,
participant_1, level_5, intensity_1_15, output_1_15
participant_1, level_5, intensity_2_15, output_2_15,
...
participant_1, level_5, intensity_100_15, output_100_15,
xxx
participant_2, level_1, intensity_1_21, output_1_21
participant_2, level_1, intensity_2_21, output_2_21,
...
participant_2, level_1, intensity_100_21, output_100_21,
participant_2, level_2, intensity_1_22, output_1_22
participant_2, level_2, intensity_2_22, output_2_22,
...
participant_2, level_2, intensity_100_22, output_100_22,
participant_2, level_3, intensity_1_23, output_1_23
participant_2, level_3, intensity_2_23, output_2_23,
...
participant_2, level_3, intensity_100_23, output_100_23,
...
I have a total of 10 participants and 6 levels. For a given (participant, level) combination, I have multiple observations which vary with intensity.
The problem is – for a given participant, I have values for only some of the levels, ie, for participant_1, I have observations for only level_1 and level_5, and for participant_2 I have observations for level_1, level_2, level_3 and there are some other participants for whom I have observations for all the levels.
So something like this would work but it would have extra ‘unused’ parameters:
(numpyro)
with numpyro.plate("n_levels", n_levels):
a_level = numpyro.sample("a_level", dist.HalfNormal(a_level_global_scale))
b_level = numpyro.sample("b_level", dist.HalfNormal(b_level_global_scale))
with numpyro.plate("n_participants", n_participants):
a = numpyro.sample("a", dist.Normal(a_level, a_global_scale))
b = numpyro.sample("b", dist.Normal(b_level, b_global_scale))
This would have 10*6 parameters for a and b.
What would be the optimal way to write down the hierarchical model without the extra parameters?
Code snippets from numpyro would help.