Hello,
I try to implement a hierarchical model with numpyro based on this tutorial. I slightly modified the tutorial. I use a quadratic model instead of a linear one and generate my own data set.
My training data is generated by the the following code snippet:
import numpy as np
import pandas as pd
def simulation_data(n: int):
res = list()
dT = np.arange(-40, 51, 5)
a = 1
sigma_a = 1
b = 2
sigma_b = 1
c = 0.5
sigma_c = 1
for idx in range(n):
alpha0_offset = np.random.normal(0, 1)
a0 = a + alpha0_offset * sigma_a
alpha1_offset = np.random.normal(0, 1)
a1 = b + alpha1_offset * sigma_b
alpha2_offset = np.random.normal(0, 1)
a2 = c + alpha2_offset * sigma_c
y_true = a0 + a1 * dT + a2 * dT**2
sigma = np.random.exponential(1)
y = np.random.normal(y_true, sigma)
res.append(pd.DataFrame({'idx' : idx, 'dT' : dT, 'y' : y}))
return pd.concat(res)
My model uses priors which are identical to the distributions above. Furthermore I use a non-centered version of the hierachical model to rule out divergences.
def model_non_centered(idx, dT, y=None):
a = numpyro.sample("a", dist.Normal(1, 0.1))
sigma_a = numpyro.sample("sigma_a", dist.Exponential(1))
b = numpyro.sample("b", dist.Normal(2., 0.1))
sigma_b = numpyro.sample("sigma_b", dist.Exponential(1))
c = numpyro.sample("c", dist.Normal(0.5, 0.1))
sigma_c = numpyro.sample("sigma_c", dist.Exponential(1))
NUM_PARTS = len(np.unique(idx))
with numpyro.plate("usnrs", NUM_PARTS):
alpha0_offset = numpyro.sample("alpha0_offset", dist.Normal(0., 1.))
alpha0 = numpyro.deterministic("alpha0", a + alpha0_offset * sigma_a)
alpha1_offset = numpyro.sample("alpha1_offset", dist.Normal(0., 1.))
alpha1 = numpyro.deterministic("alpha1", b + alpha1_offset * sigma_b)
alpha2_offset = numpyro.sample("alpha2_offset", dist.Normal(0., 1.))
alpha2 = numpyro.deterministic("alpha2", c + alpha2_offset * sigma_c)
mu = numpyro.deterministic("mu", alpha0[idx] + alpha1[idx] * dT + alpha2[idx] * dT**2)
eps = numpyro.sample("eps", dist.Exponential(1))
with numpyro.plate("data", len(idx)):
numpyro.sample("obs", dist.Normal(mu, eps), obs=y)
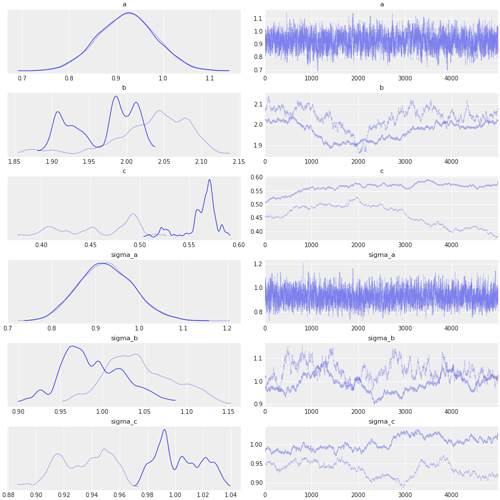
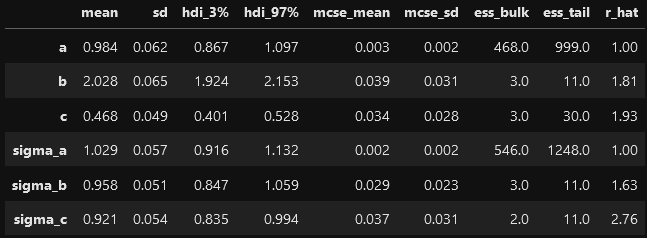
Unfortunately the model does seem to converge.
simulation = simulation_data(200)
rng_key = random.PRNGKey(1)
nuts_kernel = NUTS(model_non_centered)
non_centered_mcmc = MCMC(nuts_kernel,
num_samples=5000,
num_warmup=1000,
num_chains=2)
non_centered_mcmc.run(rng_key,
simulation.idx.values,
simulation.dT.values,
simulation.y.values)
The effective sample size of e.g. b, c, sigma_b and sigma_c is very small and MCMC does not seem to explore the parameter space appropriately.
I am new to Bayesian modeling and numpyro and I am struggling to solve this issue. Can somebody please point me into the right direction?