Hello @fehiepsi
I still do not have access to my GPU cluster, but I have set up a smaller use-case that we can dig a little bit more.
import numpy as np
import jax
import jax.numpy as jnp
from jax.config import config
config.update("jax_enable_x64", True)
import numpyro
import numpyro.distributions as dist
import numpyro.infer.autoguide as autoguide
from numpyro.infer import Predictive, SVI, Trace_ELBO, TraceMeanField_ELBO
from numpyro.optim import Adam
from numpyro.infer.reparam import NeuTraReparam
from numpyro.handlers import seed, trace, condition
from numpyro.infer import MCMC, NUTS, init_to_sample
mpl.rc('image', cmap='jet')
mpl.rcParams['font.size'] = 18
mpl.rcParams["font.family"] = "Times New Roman"
import corner
import matplotlib as mpl
import matplotlib.pyplot as plt
#For contour ploting
#-----------------------
def plot_diagnostics(samples, param_true, samples_b=None, labels=None):
mpl.rcParams['font.size'] = 12
#DF ndim = len(samples.keys())
ndim = samples.shape[1]
# This is the empirical mean of the sample:
##DF value2 = np.mean(np.array(list(samples.values())),axis=1)
value2 = np.mean(samples,axis=0)
#True
value1 = param_true
# Make the base corner plot
# 68% et 95% quantiles 1D et levels in 2D
figure = corner.corner(samples,labels=labels,quantiles=(0.025, 0.158655, 0.841345, 0.975), levels=(0.68,0.95),
show_titles=True, title_kwargs={"fontsize": 14},
truths=param_true, truth_color='g', color='b'
);
if samples_b is not None:
corner.corner(samples_b,labels=labels,quantiles=(0.025, 0.158655, 0.841345, 0.975), levels=(0.68,0.95),
show_titles=True, title_kwargs={"fontsize": 14},
truths=param_true, truth_color='g', color='purple', fig=figure
);
# Extract the axes
axes = np.array(figure.axes).reshape((ndim, ndim))
# Loop over the diagonal
for i in range(ndim):
ax = axes[i, i]
ax.axvline(value2[i], color="r")
# Loop over the histograms
for idy in range(ndim):
for idx in range(idy):
ax = axes[idy, idx]
ax.axvline(value2[idx], color="r")
ax.axhline(value2[idy], color="r")
ax.plot(value2[idx], value2[idy], "sr")
return figure
####
# Mock data
####
param_true = np.array([1.0, 0.0, 0.2, 0.5, 1.5])
sample_size = 5_000
sigma_e = param_true[4] # true value of parameter error sigma
random_num_generator = np.random.RandomState(0)
xi = 5*random_num_generator.rand(sample_size)-2.5
e = random_num_generator.normal(0, sigma_e, sample_size)
#e = np.zeros(sample_size)
yi = param_true[0] + param_true[1] * xi + param_true[2] * xi**2 + param_true[3] *xi**3# + e
plt.hist2d(xi, yi, bins=50);
###
# Simple Numpyro model
###
def my_model(Xspls,Yspls=None):
a0 = numpyro.sample('a0', dist.Normal(0.,10.))
a1 = numpyro.sample('a1', dist.Normal(0.,10.))
a2 = numpyro.sample('a2', dist.Normal(0.,10.))
a3 = numpyro.sample('a3', dist.Normal(0.,10.))
mu = a0 + a1*Xspls + a2*Xspls**2 + a3*Xspls**3
return numpyro.sample('obs', dist.Normal(mu, sigma_e), obs=Yspls)
###
# Use NUTS sampling to get a reference
###
# Start from this source of randomness. We will split keys for subsequent operations.
rng_key = jax.random.PRNGKey(0)
_, rng_key, rng_key1, rng_key2 = jax.random.split(rng_key, 4)
# Run NUTS.
kernel = NUTS(my_model, init_strategy=numpyro.infer.init_to_median())
num_samples = 10_000
n_chains = 1
mcmc = MCMC(kernel, num_warmup=1_000, num_samples=num_samples,
num_chains=n_chains,progress_bar=True)
mcmc.run(rng_key, Xspls=xi, Yspls=yi)
mcmc.print_summary()
samples_nuts = mcmc.get_samples()
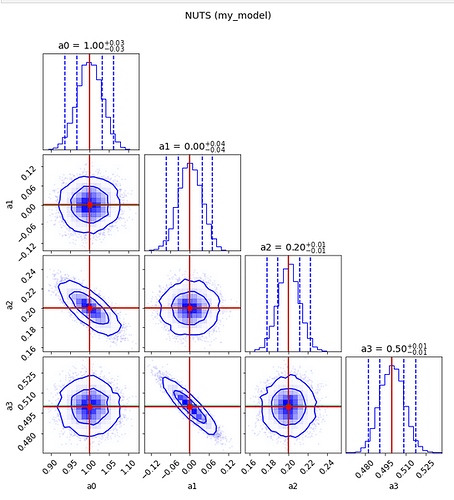
The result looks ok
mean std median 5.0% 95.0% n_eff r_hat
a0 1.00 0.03 1.00 0.95 1.05 4244.95 1.00
a1 0.00 0.04 0.00 -0.06 0.06 5130.94 1.00
a2 0.20 0.01 0.20 0.18 0.22 4405.43 1.00
a3 0.50 0.01 0.50 0.48 0.51 5174.54 1.00
Number of divergences: 0
Do the contour plots
labels = [*samples_nuts]
values = np.array(list(samples_nuts.values())).T
fig = plot_diagnostics(values, param_true[:-1], labels=labels)
fig.suptitle("NUTS (my_model)",y=1.05);
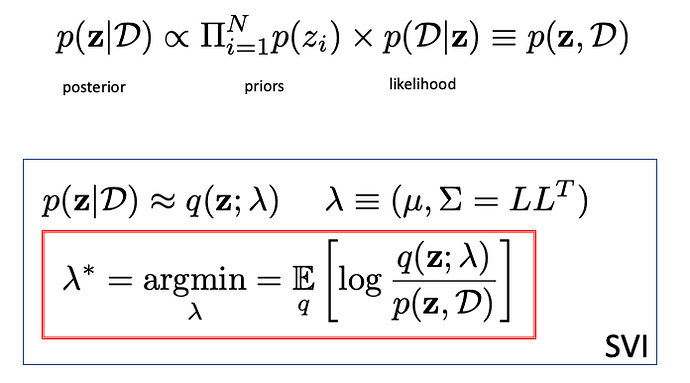
Now perform a SVI with MVN guide
guide = autoguide.AutoMultivariateNormal(my_model, init_loc_fn=numpyro.infer.init_to_sample())
optimizer = numpyro.optim.Adam(step_size=5e-3)
svi = SVI(my_model, guide,optimizer,loss=Trace_ELBO())
svi_result = svi.run(jax.random.PRNGKey(0), 10000, Xspls=xi, Yspls=yi)
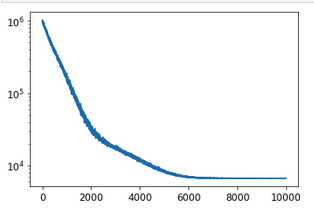
The loss decreases rather well
plt.plot(svi_result.losses)
plt.yscale('log')
Now perform the sampling of the optimised guide
samples_svi = guide.sample_posterior(jax.random.PRNGKey(1), svi_result.params, sample_shape=(5000,))
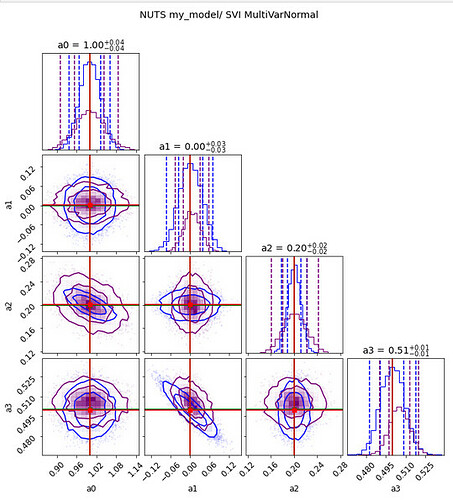
As you see below here is the comparison of the two set of contour plots
labels = [*samples_nuts]
values1 = np.array(list(samples_nuts.values())).T
values2 = np.array(list(samples_svi.values())).T
fig = plot_diagnostics(values1, param_true[:-1], samples_b=values2, labels=labels)
fig.suptitle("NUTS my_model/ SVI MultiVarNormal",y=1.05);
The blue contours are those of NUTS(my_model) and the others from the SVI optimized model
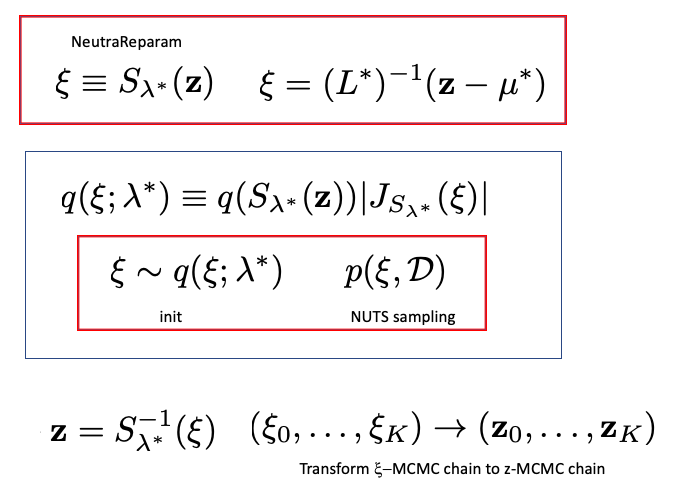
Now the Stnadard NeutraParam would be
from numpyro.infer.reparam import NeuTraReparam
neutra = NeuTraReparam(guide, svi_result.params)
neutra_model = neutra.reparam(my_model)
And one can perform a NUTS sampling based on this neutra_model as
# NUTS
####
nuts_kernel = NUTS(neutra_model)
mcmc_neutra = MCMC(nuts_kernel, num_warmup=1_000, num_samples=num_samples,
num_chains=n_chains,progress_bar=True)
mcmc_neutra.run(rng_key, Xspls=xi, Yspls=yi)
mcmc_neutra.print_summary()
####
#Get the MCMC chain with the original latent variables
zs = mcmc_neutra.get_samples()["auto_shared_latent"]
samples_nuts_neutra = neutra.transform_sample(zs)
#Compare the contours
labels = [*samples_nuts]
values1 = np.array(list(samples_nuts.values())).T
values2 = np.array(list(samples_nuts_neutra.values())).T
fig = plot_diagnostics(values1, param_true[:-1], samples_b=values2, labels=labels)
fig.suptitle("NUTS my_model/ NUTS neutra MVN",y=1.05);
And one would be happy.
Well so far so good in the context of this simple exercice everything is rather ok. But already one can question the SVI optimized solution has the sampling of this solution leads to contour plots that barely fit the true solution. Notably we do not get as correlated features than one expects. So, for my more complex example, the SVI optimized solution looks very much as Normal Independent Gaussian priors. So, it is why we engaged the discussion on a new modelling.
I have not yet manage to get it right, and I need a little help. Looking at svi_results yields
SVIRunResult(params={'auto_loc': DeviceArray([0.99782024, 0.00510312, 0.20099757, 0.50513794], dtype=float64), 'auto_scale_tril': DeviceArray([[ 4.38094511e-02, 0.00000000e+00, 0.00000000e+00,
0.00000000e+00],
[ 3.83301696e-04, 2.65209844e-02, 0.00000000e+00,
0.00000000e+00],
[-8.11400370e-03, -1.22554246e-05, 1.90036797e-02,
0.00000000e+00],
[-4.52685904e-05, -4.21003102e-03, 6.09038196e-05,
6.50602519e-03]], dtype=float64)}, state=SVIState(optim_state=(DeviceArray(10000, dtype=int64, weak_type=True), OptimizerState(packed_state=([DeviceArray([0.99782024, 0.00510312, 0.20099757, 0.50513794], dtype=float64), DeviceArray([ 16.40012248, -15.07899987, 39.20599832, -20.59373367], dtype=float64), DeviceArray([ 254334.14146581, 49334.38058495, 1307344.81640189,
893760.92674789], dtype=float64)], [DeviceArray([ 1.44527703e-02, -4.26970136e-01, -6.44897449e-04,
-6.95794883e-03, -6.47097252e-01, 9.36114107e-03,
-3.10592101e+00, -3.61652920e+00, -3.95360576e+00,
-5.03177180e+00], dtype=float64), DeviceArray([-0.6765813 , -1.07499804, 1.19968273, -0.60765637,
0.50645236, 0.15576062, 0.20232971, 0.29539797,
6.6081198 , 2.87223864], dtype=float64), DeviceArray([ 207.87352414, 12813.09351795, 13165.76001714,
2787.26121383, 2849.72120231, 3101.19997409,
2239.0793659 , 201.43236727, 11896.08727525,
2447.8603139 ], dtype=float64)]), tree_def=PyTreeDef({'auto_loc': *, 'auto_scale_tril': *}), subtree_defs=(PyTreeDef((*, *, *)), PyTreeDef((*, *, *))))), mutable_state=None, rng_key=DeviceArray([1267660082, 1240493033], dtype=uint32)), losses=DeviceArray([1017751.28605802, 988035.41332936, 1001172.51960345, ...,
6660.3973429 , 6648.90343705, 6646.57863752], dtype=float64))
How how I can complete this new model and use it concretly with the material of my simple example?
def new_model(data):
loc = numpyro.sample("loc", dist.Cauchy(0.,10.))
concentration = jnp.ones(1)
d = svi_result.params['auto_scale_tril'].shape[0]
corr_cholesky = numpyro.sample("corr_cholesky", dist.LKJCholesky(d,concentration))
scale = numpyro.sample("scale", Exponential(...))
scale_tril = corr_cholesky * scale[..., None]
# check svi_result.params or svi.get_params(state) for correct keys
params = {"auto_loc": loc, "auto_scale_tril": scale_tril}
neutra = NeuTraReparam(guide, params)
neutra_model = neutra.reparam(my_model)
return neutra_model(data)