Hello, I just tried the example, Example: Predator-Prey Model, with adapt_step_size=False and num_chains=2 .
For num_chains=1, adapt_step_size=True works perfectly, but num_chains > 2, MCMC with adapt_step_size=True is peding forever, so I changed it to False. Also, I added the lines to enable multiple draws on the CPU.
numpyro.set_host_device_count(8)
print(jax.local_device_count())
When I plot the trace of sigma, weird patterns are shown as follows
sigma = mcmc.get_samples()["sigma"]
# The number of samples = 1000.
plt.plot(sigma[:, 0])
plt.plot(sigma[:, 1])
plt.show()
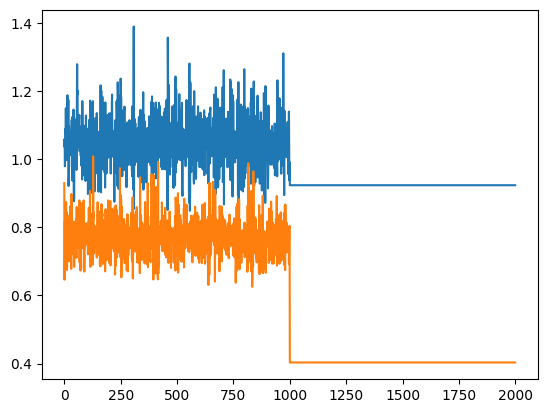
Modification of ode options such as rtol , atol , maxsteps dosen’t solve the problem. As far as I know, num_chains = 2 just performs sampling twice with difference random states (or starting potins for params maybe?). But It seems the second draw wasn’t performed. Any suggestions would be helpful.
Thank you.