I’m new to NumPyro, and I’m learning it with an aim to replace my current Stan model with Numpyro and achieve potential speedup.
I wrote a simple Gaussian Mixture Model - based on the model here, but with an added constraint that the cluster means be ordered. I’m testing it using a dataset of N = 100000 points, K = 4 clusters.
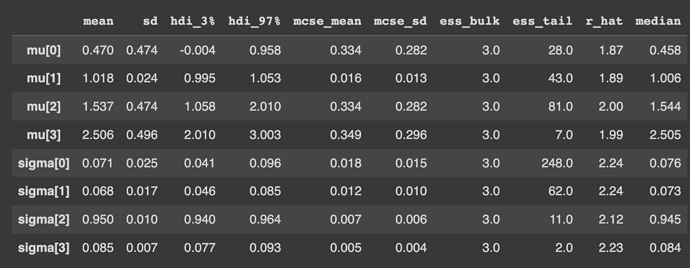
The sampler runs in ~20 minutes when using 2 chains and 1000 samples + 1000 warmups per chain, which is really fast! However, the effective sample size ess_bulk is only 3 for the means and std-devs of the clusters, which is extremely low.
I’ve created a notebook with my model, data and results - Gaussian Mixture Model - Google Colab
Also sharing here for convenience
Model
def gmm_model(y, K):
#mixture component probabilities
lambdas = numpyro.sample("lambdas", dist.Dirichlet(concentration=jnp.ones(K)))
#mu is vector of cluster means, should be an order vector drawn from a Normal distribution.
#Using a transformed distribution with an ordered transform.
mu = numpyro.sample("mu", dist.TransformedDistribution(dist.Normal(0, 10).expand([K]), transforms.OrderedTransform()))
#I'm guessing this is equivalent to expand()? Maybe it does stuff in parallel, but expand should do that too?
with numpyro.plate("K", K):
#sigma is vector of cluster stddevs
sigma = numpyro.sample("sigma", dist.HalfNormal(scale=0.1))
with numpyro.plate("N", len(y)):
#cluster_idx is vector of index to which cluster a data point belongs to.
#add infer={'enumerate': 'parallel'} to tell MCMC to marginalize discrete variable.
cluster_idx = numpyro.sample("cluster_idx", dist.Categorical(lambdas), infer={'enumerate': 'parallel'})
#likelihood - y ~ normal(mu[c], sigma[c])
numpyro.sample("y", dist.Normal(loc=mu[cluster_idx], scale=sigma[cluster_idx]), obs=y)
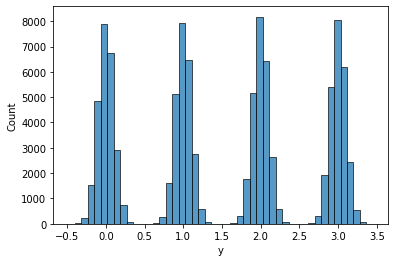
Here’s what the data looks like
And here is the summary, which includes the effective sample size ess_bulk.
I’d appreciate any advice on how I can fix this problem: is my model misspecified, or do I just need to run with more samples?