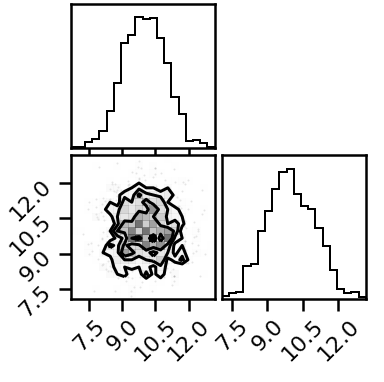
Hi @fehlespi!
I’ve been hacking away at this today with no luck, but I’ve figured some things out. Both the vanilla HMC sampler and the NeuTra HMC sampler (as used in that tutorial) seem to both struggle with sampling bimodal distributions. Perhaps this is a problem with initial guesses. See code below.
I was wondering whether a Nested Sampler would be the way to go instead, as I hear they’re better at drawing from multimodal distributions. However following this example (Example: Nested Sampling for Gaussian Shells — NumPyro documentation) results in a NotImplementedError for me, so wasn’t sure if the method was fully functional yet in Numpyro.
class mixmodel_2D(dist.Distribution):
support = constraints.real_vector
def __init__(self):
super(mixmodel_2D, self).__init__(event_shape=(2,))
def sample(self, key, sample_shape=()):
return jnp.zeros(
sample_shape + self.shape()
) # a dummy sample to initialize the samplers
# Multivariate normal distribution, takes means and covariance
def gauss_mv(self, x, locs, covs):
d = len(locs)
xm = x - locs
A = (jnp.dot(jnp.dot(xm, jnp.linalg.inv(covs)), xm.T))
B = d*jnp.log(2*jnp.pi) + jnp.log(jnp.linalg.det(covs))
return -0.5 * (A+B)
def log_prob(self, x):
locs = np.array([[-15, 10],
[-15, 20],
[-5, 15]])
cov = np.array([[5, 0],
[0, 5]])
covs = jnp.array([cov, cov, cov])
weights = jnp.array([0.1, 0.3, 0.6])
terms = jnp.array([self.gauss_mv(x, locs[i], covs[i]) for i in range(len(locs))])
args = jnp.array([terms[i] + jnp.log(weights[i]) for i in range(len(locs))])
ll = jnp.sum(jnp.log(jnp.sum(jnp.array([jnp.exp(args[i]) for i in range(len(locs))]),axis=0)))
return ll
def model():
npy.sample('y', mixmodel_2D())
num_warmup = 100
num_samples = 1000
num_chains = 1
num_iters = 1000
## First a regular NUTS HMC
nuts_kernel = NUTS(model)
mcmc = MCMC(
nuts_kernel,
num_warmup=num_warmup,
num_samples=num_samples,
num_chains=num_chains,
)
mcmc.run(key)
mcmc.print_summary()
vanilla_samples = mcmc.get_samples()["y"].copy()
#Now the NeuTra HMC
guide = AutoBNAFNormal(
model, hidden_factors=[8, 8]
)
svi = SVI(model, guide, optim.Adam(0.003), Trace_ELBO())
print("Start training guide...")
svi_result = svi.run(key, num_iters)
print("Finish training guide. Extract samples...")
guide_samples = guide.sample_posterior(
random.PRNGKey(2), svi_result.params, sample_shape=(num_samples,)
)["y"].copy()
print("\nStart NeuTra HMC...")
neutra = NeuTraReparam(guide, svi_result.params)
neutra_model = neutra.reparam(model)
nuts_kernel = NUTS(neutra_model)
mcmc = MCMC(
nuts_kernel,
num_warmup=num_warmup,
num_samples=num_samples,
num_chains=num_chains,
)
mcmc.run(random.PRNGKey(3))
mcmc.print_summary()
zs = mcmc.get_samples(group_by_chain=True)["auto_shared_latent"]
print("Transform samples into unwarped space...")
samples = neutra.transform_sample(zs)
print_summary(samples)
zs = zs.reshape(-1, 2)
samples = samples["y"].reshape(-1, 2).copy()
In the figures below, the red dots indicate the locations of the locs for the three component gaussians in the distribution - the NeuTraHMC sampler doesn’t touch them at all.