Hello,
The following model works:
def BQR_SSVS(tau=0.5, a=5, b=.04, beta_1=0.5, beta_2=0.5, gamma_1=3, gamma_2=100, X=None, y=None):
T, K = X.shape
# Deterministic
theta = (1-2*tau)/(tau*(1-tau))
tau_star_squared = 2/(tau*(1-tau))
c = 10**(-5)
# Non-Beta Priors
sigma = numpyro.sample('sigma', dist.InverseGamma(a,1/b))
# Beta Priors
beta0 = numpyro.sample('beta0', dist.Normal(0, 1))
pi_0 = numpyro.sample('pi_0', dist.Beta(beta_1,beta_2))
with numpyro.plate("plate_beta", K):
gamma = numpyro.sample('gamma', dist.Bernoulli(pi_0))
delta = numpyro.sample('delta', dist.InverseGamma(gamma_1,gamma_2))
sigma_beta = jnp.sqrt((1-gamma)*c*delta + gamma*delta)
beta = numpyro.sample("beta", dist.Normal(0, sigma_beta))
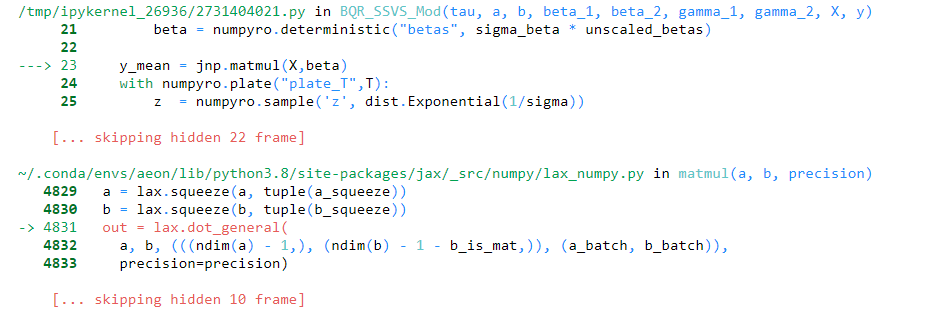
y_mean = beta0+jnp.matmul(X,beta)
with numpyro.plate("plate_T",T):
z = numpyro.sample('z', dist.Exponential(1/sigma))
sigma_obs = jnp.sqrt(tau_star_squared*sigma*z)
y = numpyro.sample("y", dist.Normal(y_mean+theta*z, sigma_obs),obs=y)
but when changing
beta = numpyro.sample("beta", dist.Normal(0, sigma_beta))
to
unscaled_betas = numpyro.sample("unscaled_betas", dist.Normal(0.0, 1.0))
beta = numpyro.deterministic("betas", sigma_beta * unscaled_betas)
It throws the following error:
It seems like changing this line changes the dims of beta? But, why?