Hello,
The question of the good convergence of the SVI arises looking at a simple exercice. Here is the complete snippet:
import jax
import jax.numpy as jnp
from jax.config import config
config.update("jax_enable_x64", True)
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
mpl.rc('image', cmap='jet')
mpl.rcParams['font.size'] = 18
mpl.rcParams["font.family"] = "Times New Roman"
import corner
import arviz as az
import numpyro
from numpyro.diagnostics import hpdi
import numpyro.distributions as dist
from numpyro import handlers
from numpyro.infer import MCMC, NUTS, init_to_sample
numpyro.util.enable_x64()
# Get mock data
param_true = np.array([1.0, 0.0, 0.2, 0.5, 1.5])
sample_size = 5_000
sigma_e = param_true[4] # true value of parameter error sigma
random_num_generator = np.random.RandomState(0)
xi = 5*random_num_generator.rand(sample_size)-2.5
# do not produce noisy data to get theoretical contours
yi = param_true[0] + param_true[1] * xi + param_true[2] * xi**2 + param_true[3] *xi**3
plt.hist2d(xi, yi, bins=50);
####
# Build Numpyro model
####
def my_model(Xspls,Yspls=None):
a0 = numpyro.sample('a0', dist.Normal(0.,10.))
a1 = numpyro.sample('a1', dist.Normal(0.,10.))
a2 = numpyro.sample('a2', dist.Normal(0.,10.))
a3 = numpyro.sample('a3', dist.Normal(0.,10.))
mu = a0 + a1*Xspls + a2*Xspls**2 + a3*Xspls**3
return numpyro.sample('obs', dist.Normal(mu, sigma_e), obs=Yspls)
###
# NUTS sampling to compare with the Truth and the SVI sampling
###
# Start from this source of randomness. We will split keys for subsequent operations.
rng_key = jax.random.PRNGKey(0)
_, rng_key, rng_key1, rng_key2 = jax.random.split(rng_key, 4)
# Run NUTS.
kernel = NUTS(my_model, init_strategy=numpyro.infer.init_to_median())
num_samples = 10_000
n_chains = 1
mcmc = MCMC(kernel, num_warmup=1_000, num_samples=num_samples,
num_chains=n_chains,progress_bar=True)
mcmc.run(rng_key, Xspls=xi, Yspls=yi)
mcmc.print_summary()
samples_nuts = mcmc.get_samples()
#####
# SVI
#####
import numpyro.infer.autoguide as autoguide
from numpyro.infer import Predictive, SVI, Trace_ELBO, TraceMeanField_ELBO
from numpyro.optim import Adam
guide = autoguide.AutoMultivariateNormal(my_model, init_loc_fn=numpyro.infer.init_to_sample())
optimizer = numpyro.optim.Adam(step_size=5e-3)
svi = SVI(my_model, guide,optimizer,loss=Trace_ELBO())
svi_result = svi.run(jax.random.PRNGKey(0), 10000, Xspls=xi, Yspls=yi)
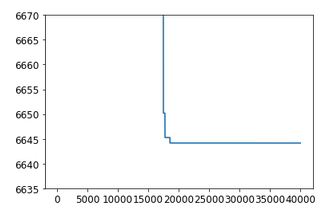
Now, ploting the loss
plt.plot(svi_result.losses)
plt.yscale('log')
gets a reasonable behaviour
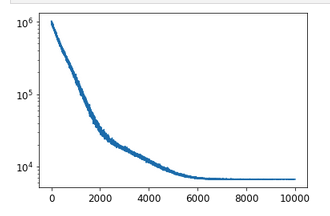
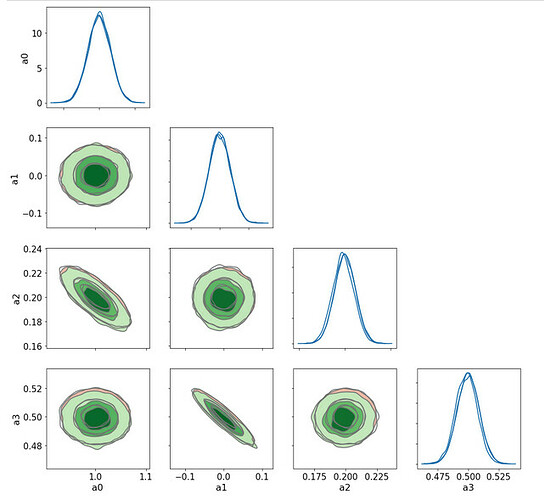
So one gets samples from SVI optimized guide
samples_svi = guide.sample_posterior(jax.random.PRNGKey(1), svi_result.params, sample_shape=(5000,))
What about the result of the linear least square fit
Y = yi[:,np.newaxis] # output (N,1)
X = np.vstack((np.ones(sample_size),xi,xi**2,xi**3)).T # input features (N,4)
K = X.T @ X
Theta_hat= (np.linalg.inv(K) @ X.T @ Y).squeeze() # fitted theta vector Y=X @ Theta
Sigma_theta = sigma_e**2 * np.linalg.inv(K) # Covariance of Theta
One gets Theta_hat = true vector
array([1.00000000e+00, 2.52575738e-15, 2.00000000e-01, 5.00000000e-01])
Get samples from MVN with loc=Theta_hat and cov mtx = Sigma_theta
dist_theta = dist.MultivariateNormal(loc=Theta_hat, covariance_matrix=Sigma_theta)
samples_true = dist_theta.sample(jax.random.PRNGKey(42), (100_000,))
spl_true = {labels[i]:samples_true[:,i] for i in range(len(labels))}
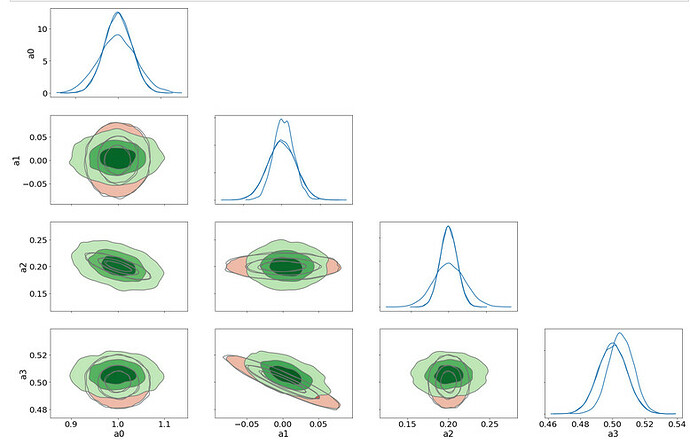
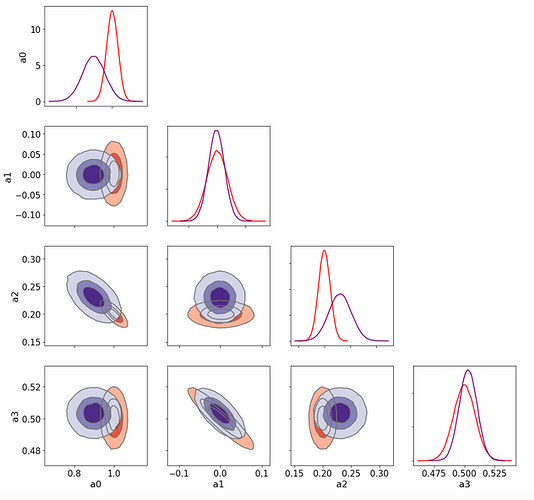
Now plot 1D and 2D Kde plots of
spl_true: a sampling from the LSQ fit in red which leads to MVN distrib
samples_nuts: the NUTS sampling of the model in blue
samples_svi: The SVI sampling from a MVN guide optimized on model in green
labels = [*samples_nuts]
ax = az.plot_pair(
spl_true,
kind="kde",
var_names=labels,
kde_kwargs={
"hdi_probs": [0.3, 0.6, 0.9], # Plot 30%, 60% and 90% HDI contours
"contourf_kwargs": {"cmap": "Reds"},
},
marginals=True, textsize=20,
);
az.plot_pair(
samples_nuts,
kind="kde",
var_names=labels,
kde_kwargs={
"hdi_probs": [0.3, 0.6, 0.9], # Plot 30%, 60% and 90% HDI contours
"contourf_kwargs": {"cmap": "Blues", "alpha": 0.2},
"plot_kwargs":{"color":"blue"}
},
marginals=True, textsize=20,ax=ax
);
az.plot_pair(
samples_svi,
kind="kde",
var_names=labels,
kde_kwargs={
"hdi_probs": [0.3, 0.6, 0.9], # Plot 30%, 60% and 90% HDI contours
"contourf_kwargs": {"cmap": "Greens", "alpha": 1.0},
"plot_kwargs":{"color":"green"}
},
marginals=True, textsize=20,ax=ax
);
Here is the result
The NUTS sampling agrees very well with the Lsq Fit ones (the two sets of contours 1D/2D are barely distinguishable)
But the SVI (green) contours (and 1D pdf too) are quite different and I do not understand why there are such as the true posterior pdf is a MVN so it is the same distribution family as the guide distrib. So, is there a problem of convergence even in this 4-parameters very simple exercice?