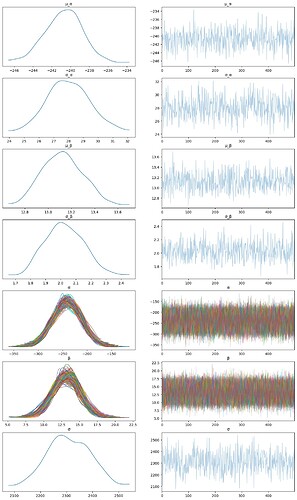
Hi all,
I try to run the bayesian hierarchical linear regression tutorial as Bayesian Hierarchical Linear Regression — NumPyro documentation and got the same results with tutorial.
But when I try to use SVI to solve the same problem, I got total different results. Did anything go wrong? Any suggestion will be useful.
Best !
Below is full codes for SVI
import torch
import pandas as pd
import numpy as np
from jax import random
from sklearn.preprocessing import LabelEncoder
import seaborn as sns
import matplotlib.pyplot as plt
import arviz as az
import pyro
import pyro.distributions as dist
from pyro.infer import SVI, Trace_ELBO, Predictive
from pyro.infer.autoguide import AutoDiagonalNormal
train = pd.read_csv(
"https://gist.githubusercontent.com/ucals/"
"2cf9d101992cb1b78c2cdd6e3bac6a4b/raw/"
"43034c39052dcf97d4b894d2ec1bc3f90f3623d9/"
"osic_pulmonary_fibrosis.csv"
)
train.head()
patient_encoder = LabelEncoder()
train["patient_code"] = patient_encoder.fit_transform(train["Patient"].values)
FVC_obs = train["FVC"].values
Weeks = train["Weeks"].values
patient_code = train["patient_code"].values
def model_pyro(patient_code, Weeks, FVC_obs=None):
μ_α = pyro.sample("μ_α", dist.Normal(0.0, 500.0))
σ_α = pyro.sample("σ_α", dist.HalfNormal(100.0))
μ_β = pyro.sample("μ_β", dist.Normal(0.0, 3.0))
σ_β = pyro.sample("σ_β", dist.HalfNormal(3.0))
n_patients = len(np.unique(patient_code))
with pyro.plate("plate_i", n_patients):
α = pyro.sample("α", dist.Normal(μ_α, σ_α))
β = pyro.sample("β", dist.Normal(μ_β, σ_β))
σ = pyro.sample("σ", dist.HalfNormal(100.0))
FVC_est = α[patient_code] + β[patient_code] * Weeks
with pyro.plate("data", len(patient_code)):
pyro.sample("obs", dist.Normal(FVC_est, σ), obs=FVC_obs)
guide = AutoDiagonalNormal(model_pyro)
adam = pyro.optim.Adam({"lr": 0.03})
svi = SVI(model_pyro, guide, adam, loss=Trace_ELBO())
patient_code = torch.tensor(patient_code)
FVC_obs = torch.tensor(FVC_obs)
Weeks = torch.tensor(Weeks)
pyro.clear_param_store()
for j in range(10000):
# calculate the loss and take a gradient step
loss = svi.step(patient_code, Weeks, FVC_obs=FVC_obs)
predictive = Predictive(model_pyro, guide=guide, num_samples=500)
preds = predictive(patient_code, Weeks)
sanitized_preds = {k: v.unsqueeze(0).detach().numpy() for k, v in preds.items() if k != 'obs'}
pyro_data = az.convert_to_inference_data(sanitized_preds)
az.plot_trace(pyro_data, compact=True,figsize=(15, 25))