Hi! A bit of a beginner question here. I’m working through some examples in Statistical Rethinking and I am having trouble understanding what is happening differently when using the BetaBinomial distribution directly vs. parameterizing a Binomial distribution with a beta distribution. The example is the admissions data with gender from chapter 12 (Mixture models) and the notebook I worked from is here.
The original version with BetaBinomial is this:
# Code 12.2
UCBadmit = pd.read_csv("../data/UCBadmit.csv", sep=";")
d = UCBadmit
d["gid"] = (d["applicant.gender"] != "male").astype(int)
dat = dict(A=d.admit.values, N=d.applications.values, gid=d.gid.values)
def model(gid, N, A=None):
a = numpyro.sample("a", dist.Normal(0, 1.5).expand([2]))
phi = numpyro.sample("phi", dist.Exponential(1))
theta = numpyro.deterministic("theta", phi + 2)
pbar = expit(a[gid])
numpyro.sample("A", dist.BetaBinomial(pbar * theta, (1 - pbar) * theta, total_count=N), obs=A)
m12_1 = MCMC(NUTS(model), num_warmup=500, num_samples=500, num_chains=4)
m12_1.run(random.PRNGKey(0), **dat)
I wanted to see what would happen when breaking it down into Beta + Binomial; the only thing that changed was the last two lines of the model where I added the p and then used the Binomial directly.
dat = dict(A=d.admit.values, N=d.applications.values, gid=d.gid.values)
def model_1a(gid, N, A=None):
a = numpyro.sample("a", dist.Normal(0, 1.5).expand([2]))
phi = numpyro.sample("phi", dist.Exponential(1))
theta = numpyro.deterministic("theta", phi + 2)
pbar = expit(a[gid])
p = numpyro.sample("p", dist.Beta(pbar * theta, (1 - pbar) * theta))
numpyro.sample("A", dist.Binomial(total_count=N, probs=p), obs=A)
m12_1a = MCMC(NUTS(model_1a), num_warmup=500, num_samples=500, num_chains=4)
m12_1a.run(random.PRNGKey(0), **dat)
The print_summary results are pretty close to the same for both models (though there’s now values for p in the second).
# Code 12.3 - original verison
post = m12_1.get_samples()
post["theta"] = Predictive(m12_1.sampler.model, post)(random.PRNGKey(1), **dat)["theta"]
post["da"] = post["a"][:, 0] - post["a"][:, 1]
print_summary(post, 0.89, False)
# Updated version with beta + binomial
post_1a = m12_1a.get_samples()
post_1a["theta"] = Predictive(m12_1a.sampler.model, post_1a)(random.PRNGKey(1), **dat)["theta"]
post_1a["da"] = post_1a["a"][:, 0] - post_1a["a"][:, 1]
print_summary(post_1a, 0.89, False)
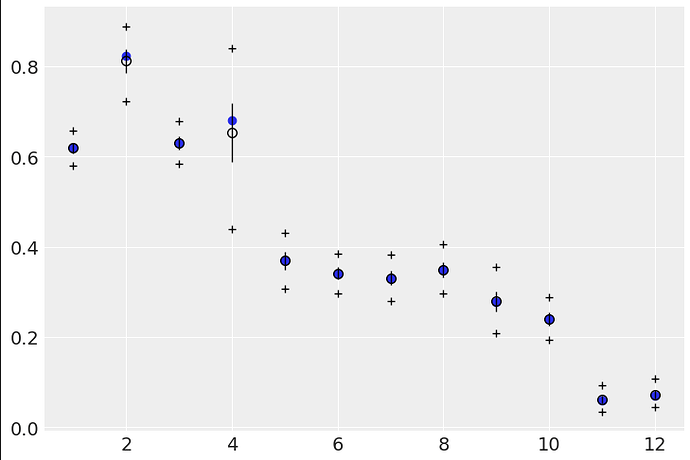
However, when doing the posterior predictive checks, the second model essentially predicts the empirical data:
Code for above:
# Code 12.5
post = m12_1a.get_samples()
admit_pred = Predictive(m12_1a.sampler.model, post_1a)(
random.PRNGKey(1), gid=dat["gid"], N=dat["N"]
)["A"]
admit_rate = admit_pred / dat["N"]
plt.scatter(range(1, 13), dat["A"] / dat["N"])
plt.errorbar(
range(1, 13),
jnp.mean(admit_rate, 0),
jnp.std(admit_rate, 0) / 2,
fmt="o",
c="k",
mfc="none",
ms=7,
elinewidth=1,
)
plt.plot(range(1, 13), jnp.percentile(admit_rate, 5.5, 0), "k+")
plt.plot(range(1, 13), jnp.percentile(admit_rate, 94.5, 0), "k+")
plt.show()
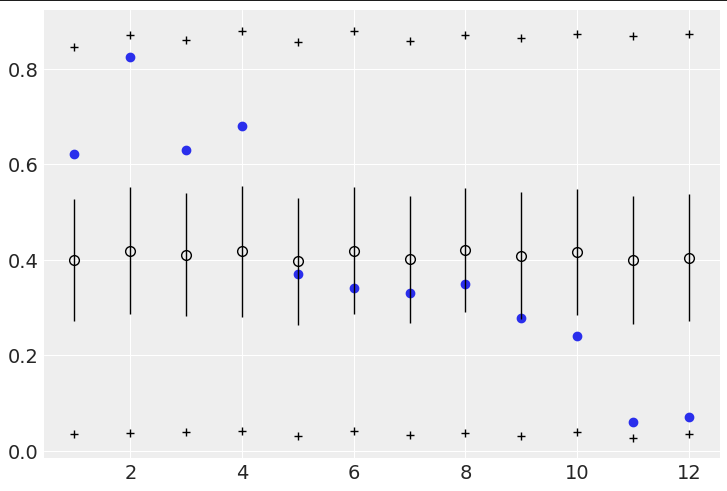
For reference, here’s the first model (code 12.5):
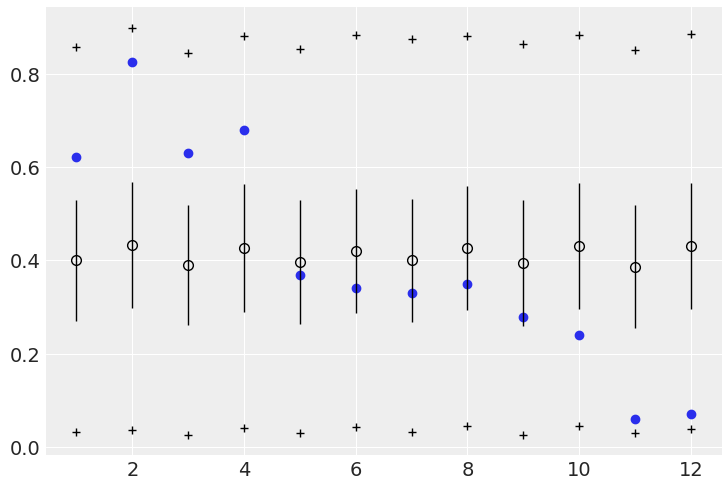
I peeked under the hood of the code for BetaBinomial and it appears to be creating a Beta distribution and sampling from it, then feeding those probs to a Binomial distribution - in other words it seems like it is doing the same thing I thought I was doing by splitting it apart.
Can anyone help me understand why there’s difference in the predicted values in this case? I think the results of the Beta + Binomial make more sense to me than the BetaBinomial, but I’m not sure how to interrogate the results to spot why it is different. Is there some kind of pooling effect happening with BetaBinomial?
Thanks, and sorry for the long post!
Ron