Wanted to provide some reproducible code. here’s the main function, I got rid of the idea of epochs for now to make it more comparable with svi.run.
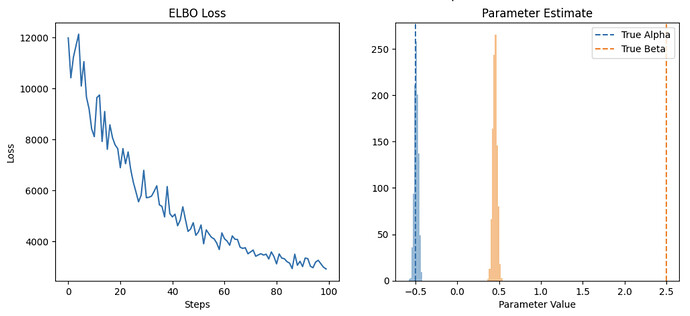
It takes a 10 seconds to fit on the data and it doesnt converge to the true parameter values after iterating through the full dataset, clearly I’m not thinking about this correctly. Is there a way to both speed this up and get it to converge to the true parameters after iterating through the full dataset?
def train_in_batches(model):
SVIRunResult = namedtuple("SVIRunResult", ("params", "state", "losses"),)
dataset= SimpleDataset(pd_dataset)
data_loder = NumpyLoader(dataset, batch_size=1000, shuffle=True)
# Define model fitting process
optimizer = numpyro.optim.Adam(0.01)
guide = AutoLowRankMultivariateNormal(model, init_loc_fn=numpyro.infer.init_to_median(), init_scale=0.025)
svi = numpyro.infer.SVI(model, guide, optimizer, Trace_ELBO())
# Set initial svi state
sample_batch = next(iter(data_loder))
svi_state = svi.init(PRNGKey(0), X=jnp.array(sample_batch[0]), y=jnp.array(sample_batch[1]))
# Train
losses = []
for x,y in tqdm(data_loder):
svi_state, loss = svi.update(svi_state, X=x, y=y)
losses.append(loss)
svi_result = SVIRunResult(svi.get_params(svi_state), svi_state, losses)
return svi_result, guide
Here’s the full code to reproduce
# ################
# Imports
# ################
from collections import namedtuple
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from torch.utils.data import Dataset
from torch.utils.data import DataLoader
from torch.utils.data import default_collate
from jax import jit, lax, random
from jax.tree_util import tree_map
import jax.numpy as jnp
from jax.random import PRNGKey
from tqdm.notebook import tqdm
import numpyro
from numpyro import optim
import numpyro.distributions as dist
from numpyro.infer import SVI, Trace_ELBO
from numpyro.infer.autoguide import AutoLowRankMultivariateNormal
# ################
# Generate Data
# ################
SEED=99
np.random.seed(SEED)
N = 100000
beta = 2.5
alpha = -0.5
X = np.random.normal(0,1,size=N)
y = alpha + beta*X + np.random.normal(size=N)
pd_dataset = pd.DataFrame({"X":X, "y":y})
# ################
# Define Model
# ################
def model(X, y=None):
beta = numpyro.sample("beta", dist.Normal(0,1))
alpha = numpyro.sample("alpha", dist.Normal(0,1))
sigma = numpyro.sample("sigma", dist.Exponential(1))
mu = alpha + beta*X
obs = numpyro.sample("obs", dist.Normal(mu, sigma), obs=y)
# #################################
# Define training procedure 1
# #################################
def train_on_all_data(model):
optimizer = numpyro.optim.Adam(0.01)
guide = AutoLowRankMultivariateNormal(
model, init_loc_fn=numpyro.infer.init_to_median(), init_scale=0.025
)
data = pd_dataset
svi = numpyro.infer.SVI(model, guide, optimizer, Trace_ELBO())
svi_result = svi.run(PRNGKey(0), 1000, X=data.X.values, y=data.y.values)
return svi_result, guide
# #################################
# summarize training results
# #################################
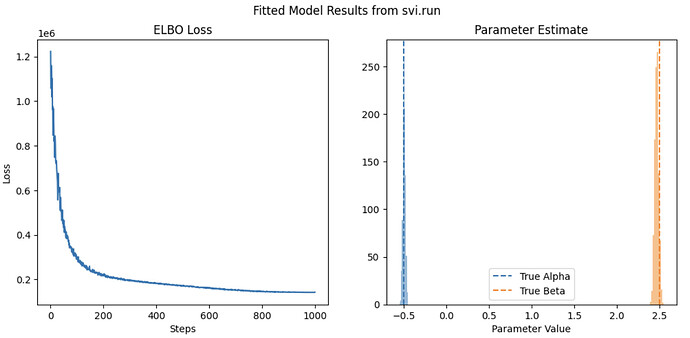
def plot_results(svi_result, guide, method='svi.run'):
fig, ax = plt.subplots(1,2, figsize=(12,5))
ax[0].set(title="ELBO Loss",xlabel='Steps', ylabel='Loss')
ax[0].plot( svi_result.losses )
ax[1].set(title="Parameter Estimate",xlabel='Parameter Value')
samples = guide.sample_posterior(PRNGKey(1), svi_result.params, (1000,))
ax[1].hist( samples['alpha'], alpha=0.5 )
ax[1].hist( samples['beta'], alpha=0.5 )
ax[1].axvline(alpha, ls='--', label='True Alpha')
ax[1].axvline(beta, ls='--', color='C1', label='True Beta')
ax[1].legend()
plt.suptitle(f"Fitted Model Results from {method}")
# #########################################################
# Create pytorch dataloader for training in batches
# #########################################################
class SimpleDataset(Dataset):
def __init__(self, df, transform=None):
self.df = df
def __len__(self):
return len(self.df)
def __getitem__(self, idx):
vals = self.df.iloc[[idx]].values
X, target = vals[:,0], vals[:,1]
return X,target
def numpy_collate(batch):
return tree_map(np.asarray, default_collate(batch))
class NumpyLoader(DataLoader):
def __init__(self, dataset, batch_size=1,
shuffle=False, sampler=None,
batch_sampler=None, num_workers=0,
pin_memory=False, drop_last=False,
timeout=0, worker_init_fn=None):
super(self.__class__, self).__init__(dataset,
batch_size=batch_size,
shuffle=shuffle,
sampler=sampler,
batch_sampler=batch_sampler,
num_workers=num_workers,
collate_fn=numpy_collate,
pin_memory=pin_memory,
drop_last=drop_last,
timeout=timeout,
worker_init_fn=worker_init_fn)
# #########################################################
# Define training procedure for training in batches
# #########################################################
def train_in_batches(model):
SVIRunResult = namedtuple("SVIRunResult", ("params", "state", "losses"),)
dataset= SimpleDataset(pd_dataset)
data_loder = NumpyLoader(dataset, batch_size=1000, shuffle=True)
# Define model fitting process
optimizer = numpyro.optim.Adam(0.01)
guide = AutoLowRankMultivariateNormal(model, init_loc_fn=numpyro.infer.init_to_median(), init_scale=0.025)
svi = numpyro.infer.SVI(model, guide, optimizer, Trace_ELBO())
# Set initial svi state
sample_batch = next(iter(data_loder))
svi_state = svi.init(PRNGKey(0), X=jnp.array(sample_batch[0]), y=jnp.array(sample_batch[1]))
# Train
losses = []
for x,y in tqdm(data_loder):
svi_state, loss = svi.update(svi_state, X=x, y=y)
losses.append(loss)
svi_result = SVIRunResult(svi.get_params(svi_state), svi_state, losses)
return svi_result, guide
svi_result, guide = train_on_all_data(model)
plot_results(svi_result, guide)
plt.show()
svi_result_batched, guide_batched = train_in_batches(model)
plot_results(svi_result_batched, guide_batched, method='batched svi.update')
plt.show()