Hello,
I am trying to combine the tutorials for the gaussian mixture model and the hidden Markov model to get a mixture of hidden Markov models but am running into some errors. The generative process is as follows:
There are K clusters, each has its own Markov model (so Kx start probs, Kx states, K x transition matrix)
For each id, choose a cluster assignment.
Given the cluster assignment, generate a sequence from the K th Markov model.
Data: num_ids c. 3000, each has 100-200 associated sequences.
Observation distribution = Normal distribution
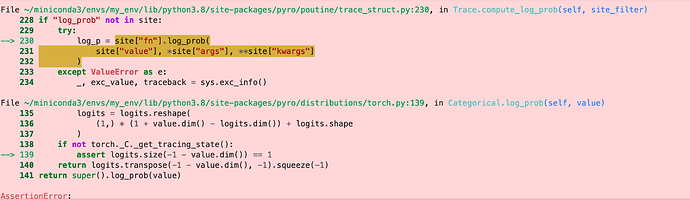
I am running into issues with batching and also with expanding the assignment tensor to all the sequences from the id - I get an assertion error at the end of an epoch due to model_trace.compute_log_prob(). I am running this on Cuda.
The model code is:
@config_enumerate
def model_7(sequences, lengths, args, batch_size=None, include_prior=True):
# Global variables.
print('idnos', idnos.shape, 'sequences', sequences.shape, 'lengths', lengths.shape)
with ignore_jit_warnings():
# Num ids, one cluster assignment per id, num_sequences per id each with max length and values of data_dim
num_ids, num_sequences, max_length, data_dim = map(int, sequences.shape)
num_clusters = args.num_clusters
# Global wvariables - mixture weights
weights = pyro.sample("probs_gmm_weights", dist.Dirichlet(torch.ones(num_clusters)/num_clusters)) #.to_event(1)
print('Draw global mixture weights', weights.shape, weights)
with pyro.plate('components', num_clusters):
# Draw start probabilities, transition matrix and means + stds for each of the components
probs_s = pyro.sample(
"probs_s",
dist.Dirichlet(torch.ones((1,args.hidden_dim))/args.hidden_dim).to_event(1),
)
probs_x = pyro.sample(
"probs_x",
dist.Dirichlet(torch.ones(args.hidden_dim, args.hidden_dim)/args.hidden_dim).to_event(1),
)
probs_y = pyro.sample(
"probs_y",
dist.Normal(loc=0, scale=1).expand([args.hidden_dim, data_dim]).to_event(2),
)
probs_z = pyro.sample(
"probs_z", dist.InverseGamma(1,0.1).expand([args.hidden_dim, data_dim]).to_event(2),
)
print('Components', probs_s.shape, probs_x.shape, probs_y.shape, probs_z.shape)
# Definte plate for observation distribution
angles_plate = pyro.plate("angles", data_dim, dim=-1)
# Draw assignments per id
assignment = pyro.sample("assignment", dist.Categorical(weights).expand([num_ids]).to_event(1))
print('Assignments for batch', assignment.shape)
with pyro.plate("sequences", num_ids*num_sequences, args.batch_size*num_sequences, dim=-2) as batch:
batch_assignments = assignment.repeat_interleave(num_sequences)[batch].squeeze()
batch_lengths = lengths.reshape(-1)[batch] # Batch by idnos
print('Batch lengths/assignments', batch_lengths.shape, batch_assignments.shape)
x=0 # Initial state 0
for t in pyro.markov(range(max_length if args.jit else batch_lengths.max())):
with handlers.mask(mask=(t < batch_lengths).unsqueeze(-1)):
if t == 0:
x = pyro.sample(
"x_{}".format(t),
dist.Categorical(probs_s[batch_assignments.squeeze(),x]),#.unsqueeze(-1) # VHANGE TO c[batch]
infer={"enumerate": "parallel"})
print("x_{}".format(t), x.shape)
else:
x = pyro.sample(
"x_{}".format(t),
dist.Categorical(probs_x[batch_assignments.squeeze(), x.squeeze(-1)]),
infer={"enumerate": "parallel"},
)
print("x_{}".format(t), x.shape)
with angles_plate:
print('Y','assignment', batch_assignments.squeeze().shape, 'rule x', x.squeeze(-1).shape,'indexed Normal means', probs_y[batch_assignments.squeeze(), x.squeeze(-1)].shape, 'observations', sequences.reshape(-1, max_length, data_dim).squeeze()[batch, t].sum())
y=pyro.sample(
"y_{}".format(t),
dist.Normal(
loc=probs_y[ batch_assignments.squeeze(), x.squeeze(-1)],
scale=probs_z[ batch_assignments.squeeze(),x.squeeze(-1)]),
obs=sequences.reshape(-1, max_length, data_dim).squeeze()[batch, t],
infer={"enumerate": "parallel"})
The guide is:
guide = AutoDelta(
handlers.block(model, expose_fn=lambda msg: msg["name"].startswith("probs_"))
)
As I said, one epoch runs fine and then I get an assertion error. I have tried various other iterations with plates around assignment etc. but all seems to either give this assertion error or a mismatch of shape inside plates due to the single assignment per id broadcast to number of sequences per id.
Does anyone know how to fix this? Any help would be much appreciated.
Thanks