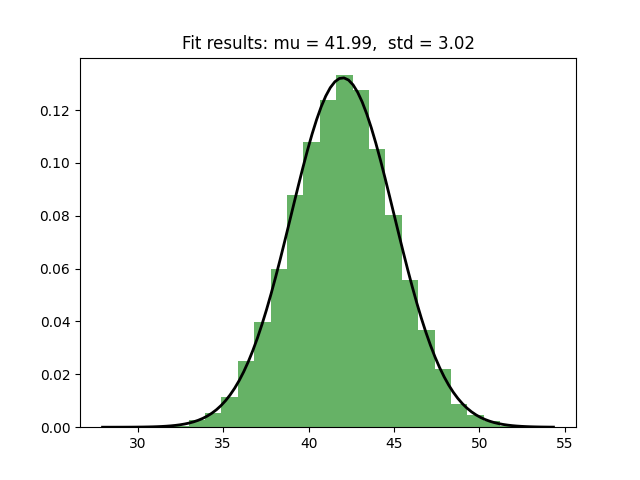
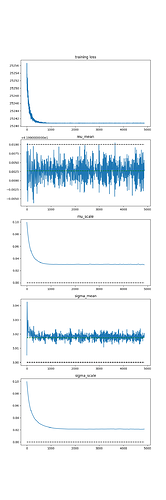
I have a conceptual problem getting Pyro to learn the parameters of a distribution using SVI. For some reason, SVI is not correctly learning the sigma/scale parameter of a normal distribution. Here is the problem setup:
%matplotlib notebook
import numpy as np
import matplotlib.pyplot as plt
from pyro.optim import Adam
from pyro.infer import SVI, Trace_ELBO
import pyro
import torch
from tqdm.notebook import tqdm
mu_prior = 42.0
sigma_prior = 1.0
def model(obs):
with pyro.plate("samples",len(obs)):
s = pyro.sample("test", dist.Normal(mu_prior,sigma_prior), obs = obs)
def guide(obs):
mu_param = pyro.param('mu_param',torch.tensor(mu_prior))
sigma_param = pyro.param('sigma_param',torch.tensor(sigma_prior))
with pyro.plate("samples",len(obs)):
s = pyro.sample("test",dist.Normal(mu_param,sigma_param))
adam_params = {"lr": 0.005, "betas": (0.95, 0.999)}
optimizer = Adam(adam_params)
svi = SVI(model, guide, optimizer, loss=Trace_ELBO(num_particles=100))
obs = torch.tensor(np.random.normal(loc=mu_prior,scale=sigma_prior,size=1000))
I would like SVI to learn mu and sigma, the parameters of the distribution. To keep things simple, I am sampling from the prior distribution to make sure that the posterior is the same as the prior. To learn the parameters, I run the following:
fig = plt.figure(figsize=(8,16))
ax1 = fig.add_subplot(311)
ax2 = fig.add_subplot(312)
ax3 = fig.add_subplot(313)
axes = ax1,ax2,ax3
plt.ion()
pyro.clear_param_store()
n_steps=20000
loss = []
mu = []
sigma = []
for step in tqdm(range(n_steps)):
l = svi.step(obs)
loss.append(l)
mu.append(pyro.param('mu_param').item())
sigma.append(pyro.param('sigma_param').item())
for ax in axes:
ax.clear()
ax1.plot(loss,label="loss")
ax1.title.set_text("training loss")
ax2.plot(mu,label="mu")
ax2.title.set_text("mu")
ax3.plot(sigma,label="sigma")
ax3.title.set_text("sigma")
fig.canvas.draw()
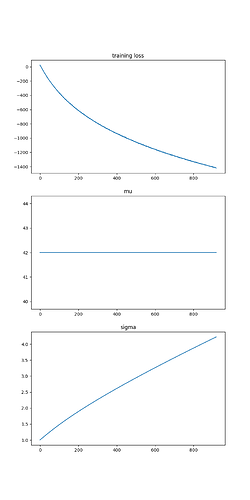
The output is:
The mean (mu) parameter is correct, but the scale parameter just continues to grow as I run SVI for more iterations. Clearly I’m missing something fundamental, but I’m at a loss. Can anyone help?