Hello again,
Related post: Batch processing Pyro models so cc: @fonnesbeck as I think he’ll be interested in batch processing Bayesian models anyway.
I want to run lots of numpyro models in parallel. I created a new post because:
- this post uses numpyro instead of pyro
- I’m doing sampling instead of SVI
- I’m using Ray instead of Dask
- that post was 2021
I’m running a simple Neal’s funnel model as a test. I have set up a small Ray cluster of 3 instances with 4CPUs each. I run the Neal’s funnel model for dim in range(2, 20). I do this once running one dim after another (sequentially in the run_model function) and once in parallel using Ray remote (in the ray_run_model decorated with @ray.remote). Full code is at the bottom.
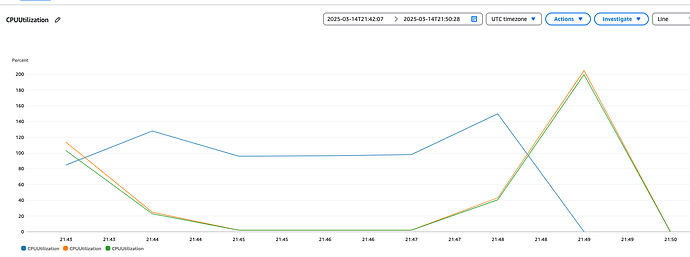
I think numpyro on Ray is working. When I look at the logs, I get a Speedup: 5.02x. And I can see the 3 instances and the CPUs being used. Below is the CPU usage plot. Spin up is until 21:45, then the sequential runs are until 21:47 and the parallel kicks in after. As the blue line is the head node, that’s the one doing running the sequential tasks and for the parallel tasks doing the scheduling across the rest of its CPUs and the CPUs on the other instances. So I’m fairly convinced it’s running a numpyro model as expected.
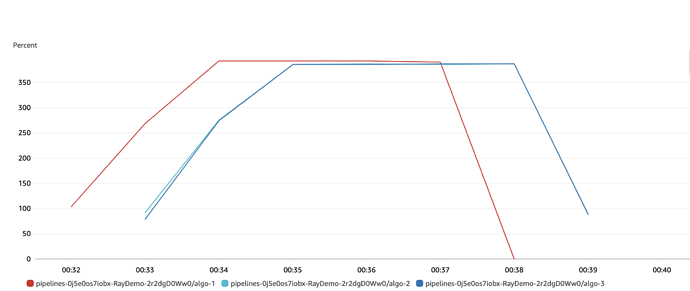
EDIT: adding a little bit of colour. I now ran the parallel test only for 1000 of the same dim models and looked at the CPU utilisation. This looks really good. In this case, the head node is in red and I made the worker nodes blues. The head node fills itself up before sending tasks to worker nodes, peaking at ~394% of the CPU on each node.
I’m experienced with numpyro but I’m unfamiliar with what goes on under the hood in XLA, and I’m inexperienced with Ray too. I have a few questions to those who might know more about how to get the most out this setup:
- In the post linked above, the dev mentioned “several pieces of global state in Pyro’s internals… I would also not be surprised to see problems with parameters from different runs overwriting each other in the global parameter store.” Is this something I should be concerned with in numpyro too?
- Further on this, to do with environment variables. I am just running chains in sequence, because I know that
numpyro.set_host_device_count(X)changes aXLA_FLAGSenvironment variable, and that this might limit how many numpyro chains can happen simultaenously on an instance. But I am interested to hear whether you think parallel chains could work anyway or if that environment variable will mess things up? (Ray might be smart enough to deal with the environment variable stuff but I have no idea.) - Although numpyro runs one chain per core, I assume there’s some multi-threading going on e.g. for matrix multiplication in XLA (or more generally in numpy) where I might use more cores. Is this the case, and if so, is it a bad idea (wrt jax) to use up all the rest of the cores for batch model runs?
- And for the Ray side of this, if I set
@ray.remote(num_cpus=2), is Ray smart enough to allocate these CPUs to the multithreading of that particular model, or are processes going to be competing with each other regardless of the resources I specify to Ray. (No worries on this if it’s not obvious as this is more a Ray question than XLA, but I’m unfamiliar with stuff so perhaps it’s obvious.)
- And for the Ray side of this, if I set
Happy to provide any more code to try things out and see how numpyro and Ray can be optimally used together. I think it’s an exciting way to scale many models/potentially chains across a cluster.
Thanks as always,
Theo
import os
import time
import jax
import jax.numpy as jnp
import numpyro
import numpyro.distributions as dist
from numpyro.infer import MCMC, NUTS
import ray
def model(dim: int = 10) -> None:
y = numpyro.sample("y", dist.Normal(0, 3))
numpyro.sample("x", dist.Normal(jnp.zeros(dim - 1), jnp.exp(y / 2)))
def run_inference(model, rng_key: jax.Array, dim: int):
kernel = NUTS(model)
mcmc = MCMC(
kernel,
num_warmup=10000,
num_samples=10000,
num_chains=2,
progress_bar=True,
)
mcmc.run(rng_key, dim=dim)
return mcmc
def run_model(dim):
rng_key = jax.random.PRNGKey(0)
mcmc = run_inference(model, rng_key, dim=dim)
samples = mcmc.get_samples()
return samples["x"].shape
@ray.remote
def ray_run_model(dim):
"""Remote version of the model with some ray variables printed"""
rng_key = jax.random.PRNGKey(0)
mcmc = run_inference(model, rng_key, dim=dim)
samples = mcmc.get_samples()
context = ray.runtime_context.get_runtime_context()
return {
"result": samples["x"].shape,
"node_id": context.get_node_id(),
"job_id": ray.get_runtime_context().get_job_id(),
"actor_id": ray.get_runtime_context().get_actor_id(),
"task_id": ray.get_runtime_context().get_task_id(),
"worker_id": ray.get_runtime_context().get_worker_id(),
"assigned_resources": ray.get_runtime_context().get_assigned_resources(),
"hostname": os.uname().nodename,
"pid": os.getpid(),
}
def test_ray_parallel():
# Test sequential execution first
print("Running sequential test...")
start_time = time.time()
sequential_results = [
run_model(dim) for dim in range(2, 20)
] # direct function calls
sequential_time = time.time() - start_time
print(f"Sequential execution took: {sequential_time:.2f} seconds")
print(f"Sequential results: {sequential_results}")
# Test parallel execution
print("\nRunning parallel test...")
start_time = time.time()
futures = [ray_run_model.remote(dim) for dim in range(2, 20)]
parallel_results = ray.get(futures) # this will run in parallel
parallel_time = time.time() - start_time
print(f"Parallel execution took: {parallel_time:.2f} seconds")
print("Parallel results: ")
for results in parallel_results:
print(results)
return sequential_time, parallel_time
if __name__ == "__main__":
ray.init(address=<>)
try:
seq_time, par_time = test_ray_parallel()
print("\nSummary:")
print(f"Sequential time: {seq_time:.2f} seconds")
print(f"Parallel time: {par_time:.2f} seconds")
print(f"Speedup: {seq_time / par_time:.2f}x")
if par_time < seq_time:
print("Ray parallel processing is working correctly!")
else:
print("Parallel execution was not faster than sequential!")
except Exception as e:
print(f"Error testing Ray: {str(e)}")
finally:
ray.shutdown()