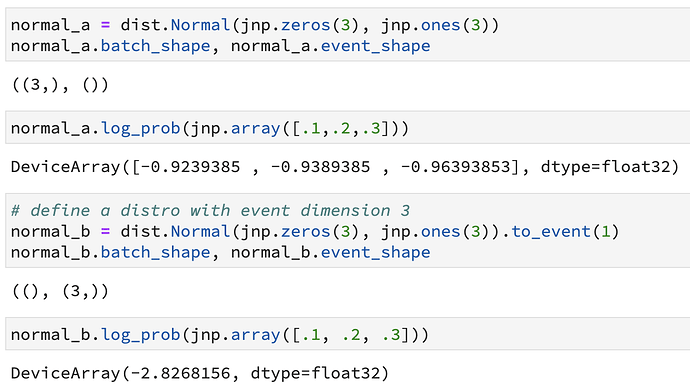
I am quite a bit confused with the way batch and event dimensions are treated when assembling the log_pdf(). As far as I understand their definitions, the event dimensions should be summed over in the log_prob(), while the batch dimensions should remain in the log_prob:
But if I do the same via a model definition, I do not observe the behaviour above. Instead, I observe this:
What confuses me: The log_pdf of the model seems to always behave like it has event dimensions. What’s going on?
More specifically:
- Why do I not see batching over independent dimensions in model_a above?
- Is there a way to examine the shapes of sites inside a model? In Pyro there is trace.format_shapes() command from this tutorial
Tensor shapes in Pyro — Pyro Tutorials 1.8.4 documentation]
Is there an equivalent way in numpyro?
Thank you for any help!
Hi @pyatsysh . I think that is because numpyro.infer.util.log_density computes log of joint density of the model, i.e. it automatically sums log probabilities over all batch dimensions.
To inspect the trace you can use numpyro.util.format_shapes (in NumPyro the trace object is just an OrderedDict).
2 Likes
Thank you for your help. I was trying to understand some of the inner workings of numpyro in dealing with batch (i.e., independent) and event (i.e., dependent) dimensions.
Specifically, I struggle to understand, how batch and event dimensions are treated differently in the posterior pdf.
Assuming, i defined the joint probability over my latent and observed variables in a typical numpyro model function. Are the following statements correct:
-
To run mcmc on the latents, an unnormalised posterior log-pdf is somehow assembled as a callable python function from the model function and the observed data
-
(!) If 1. is correct, when that unnormalised posterior log-pdf callable function is assembled, the batch and event dimensions are all summed over?
-
If 1, is correct, can i get a handle to that unnormalised posterior pdf to, e.g., evaluate it at some arbitrary values of the latents?
If 2. is correct, then there seems to be no reason to distinguish between batch and event dimensions in the unnormalised posterior log-pdf. Is that right?
I don’t know implementation details of MCMC algorithms. Maybe @fehiepsi or @martinjankowiak can provide help.
That’s right. In the context of MCMC, the notions of batch/event dimensions are not important, unless you want to do something like enumeration, where we need to vectorize the calculation over possible values of the discrete latent variables.
can i get a handle to that unnormalised posterior pdf to, e.g., evaluate it at some arbitrary values of the latents
I think you can use jax.vmap for that.
1 Like