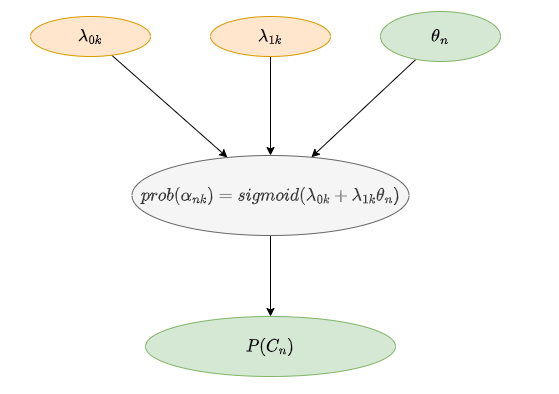
This model is related to the test. Assume there are 2 skills for everyone, and the skill master probability depends on ability \theta_n .
prob(\alpha_{nk}) =sigmod(\lambda_{0k}+\lambda_{1k}\theta_n)
\alpha_{nk} represents whether the nth person has mastered the kth skill, \theta_n is the person’s ability, and \lambda_{0k} and \lambda_{1k} represent the intercept and slope, respectively. The probability of \alpha_{nk} being 1 is as above.
All possible skill patterns can be represented as C row K col matrix, the row is skill pattern category, and the column is skill.
P=\begin{bmatrix} 0 & 0 \\1 & 0 \\0 & 1 \\ 1 & 1 \end{bmatrix}
We can get skill pattern categories by
P(C_n=c) = \sum_1^C \sum_1^KP_{ck}prob(\alpha_{nk}) +[1-P_{ck}][1-prob(\alpha_{nk})]
In my model , see skill pattern category as latent discrete variable and enumeration it. Here is my code:
N,K = 1000,4
all_p = np.array(list(itertools.product(*[[0,1] for i in range(K)])))
def M():
person_plate = numpyro.plate("person", N, dim=-2)
skill_plate = numpyro.plate("skill", K, dim=-1)
with skill_plate:
# intercept
lam0 = numpyro.sample("lam_0",dist.Normal(0, 1))
# slope
lam1 = numpyro.sample("lam_1",dist.HalfNormal(10))
with person_plate:
# ability
theta = numpyro.sample("theta", dist.Normal(0,1))
# prob(alpha_{nk})
alpha_prob = nn.sigmoid(lam0 + theta*lam1)
# transform prob(alpha_{nk}) to category_prob_{n}
category_prob = jnp.exp((jnp.log(alpha_prob).dot(all_p.T))+(jnp.log(1-alpha_prob).dot(1-all_p.T)))
category_prob = category_prob.reshape(N,1,-1)
# enumerate latent discrete variable
student_c = numpyro.sample("cat", dist.Categorical(category_prob),infer={"enumerate": "parallel"})
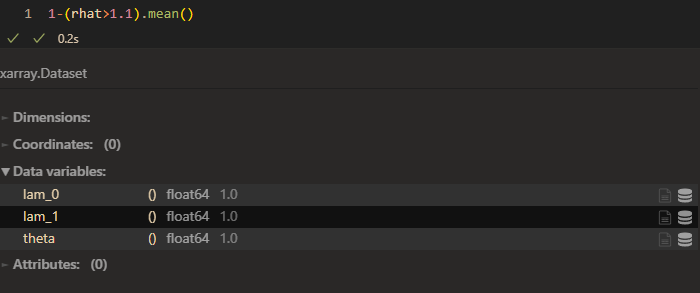
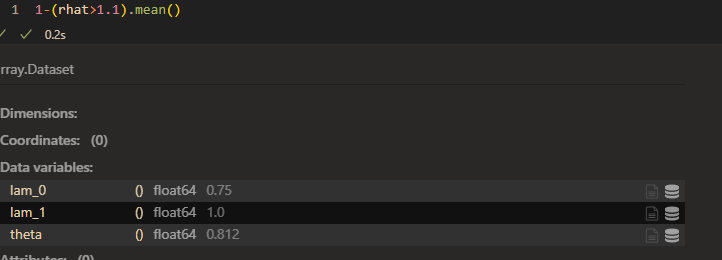
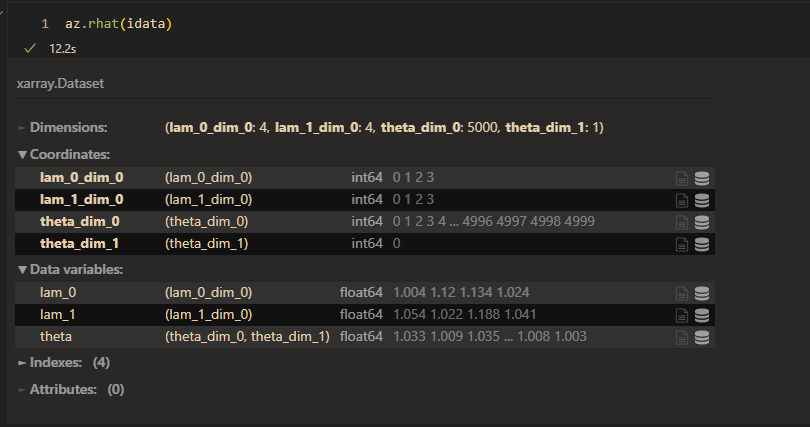
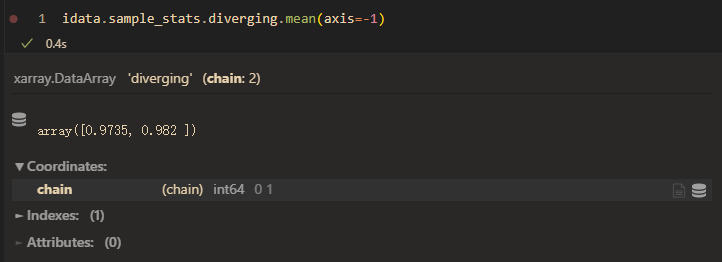
The divergence rate is high and the ess is small when I set prior of \lambda_{1k} as HalfN(0,10).
divergence mean:
ess mean:
raht:
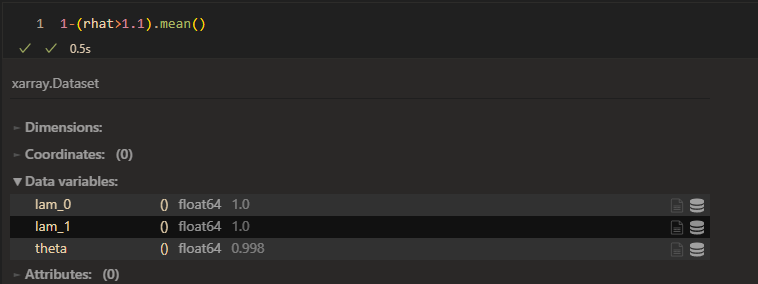
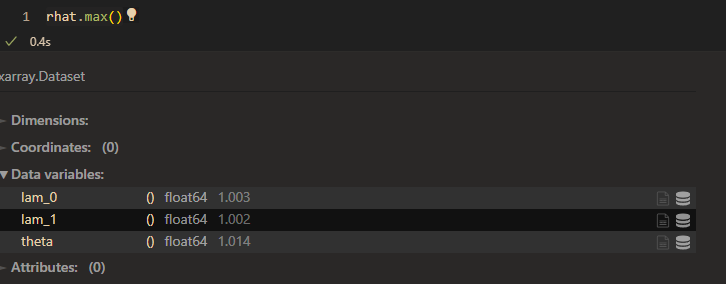
However, the model seems to work good when I set prior of \lambda_{1k} as HalfN(0,1).(small variance)
lam1 = numpyro.sample("lam_1",dist.HalfNormal(1))
divergence mean:
ess mean:
raht:
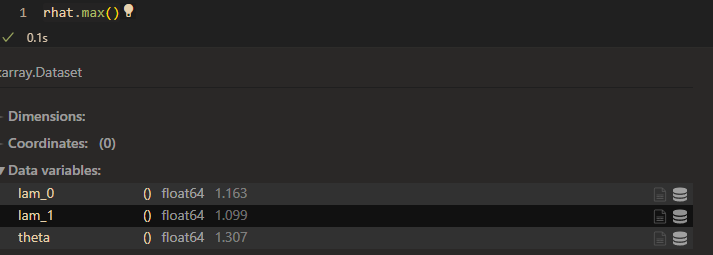
When taking more skill into consideration, the convergence is woser.
skill K = 4
N,K = 1000,4
lam1 = numpyro.sample("lam_1",dist.HalfNormal(10))
divergence mean:
ess mean:
raht:
So my question is:
- Why small variance prior work but a large variance not work?
- How can I fix it if I still want to set large variance distribution as prior for the model?
Any help would be greatly appreciated!