I’ve been at this for several days now.
I have my model defined as following:
def quadratic_model(x: List[float],
y: Optional[List[float]] = None,
weight: Union[List[float], float] = 1.0,
exercise_id: Optional[List[int]] = None,
session_id: Optional[List[int]] = None,
**kwargs) -> NoReturn:
exercise_check = exercise_id is not None
session_check = session_id is not None
# Global Parameters
global_intercept = numpyro.sample('global_intercept', dist.Normal(0.0, 1.0))
global_slope = numpyro.sample('global_slope', dist.Gamma(2.0, 1.0))
global_curvature = numpyro.sample('global_curvature', dist.HalfNormal(1.0))
# Exercise Parameters
if exercise_check:
global_intercept_scale = numpyro.sample('global_intercept_scale', dist.HalfNormal(1.0))
global_slope_rate = numpyro.sample('global_slope_rate', dist.HalfNormal(1.0))
with numpyro.plate('exercise', len(np.unique(exercise_id)), dim = -1):
exercise_intercept = numpyro.sample('exercise_intercept', dist.Normal(global_intercept, global_intercept_scale))
exercise_slope = numpyro.sample('exercise_slope', dist.Gamma(global_slope*global_slope_rate, global_slope_rate))
exercise_curvature = numpyro.sample('exercise_curvature', dist.HalfNormal(1.0/global_curvature))
# Session Parameters
if session_check:
exercise_intercept_scale = numpyro.sample('exercise_intercept_scale', dist.HalfNormal(1.0))
exercise_slope_rate = numpyro.sample('exercise_slope_rate', dist.HalfNormal(1.0))
with numpyro.plate('session', len(np.unique(session_id)), dim = -2):
session_intercept = numpyro.sample('session_intercept', dist.Normal(exercise_intercept, exercise_intercept_scale))
session_slope = numpyro.sample('session_slope', dist.Gamma(exercise_slope*exercise_slope_rate, exercise_slope_rate))
session_curvature = numpyro.sample('session_curvature', dist.HalfNormal(1.0/exercise_curvature))
# Parameter Assignments
intercept = global_intercept
slope = global_slope
curvature = global_curvature
if exercise_check:
intercept = exercise_intercept[exercise_id]
slope = exercise_slope[exercise_id]
curvature = exercise_curvature[exercise_id]
if (exercise_check) & (session_check):
intercept = session_intercept[session_id, exercise_id]
slope = session_slope[session_id, exercise_id]
curvature = session_curvature[session_id, exercise_id]
loc = intercept - slope*x - curvature*x**2
scale = numpyro.sample('error', dist.HalfNormal(1.0))
with numpyro.plate('observation', len(x)), handlers.scale(scale = weight):
numpyro.sample('y_est', dist.Normal(loc, scale), obs = y)
Using MCMC I get the following and it looks exactly like I expected:
However, when I try to use SVI with the following code, it gets nowhere near the result of MCMC:
quadratic_guide = autoguide.AutoNormal(quadratic_model, init_loc_fn = autoguide.init_to_median, init_scale = 0.01)
svi = infer.SVI(model = quadratic_model,
guide = quadratic_guide,
optim = optim.Adam(step_size = 0.01),
loss = infer.Trace_ELBO())
num_samples = 1000
svi_result = svi.run(rng_keys[2], num_samples, **data)
svi_posterior_predictive_fn = infer.Predictive(model = quadratic_model,
guide = quadratic_guide,
num_samples = num_samples,
batch_ndims = 2)
svi_posterior_predictive = svi_posterior_predictive_fn(rng_keys[5], **data)
predictions = svi_posterior_predictive_fn(rng_keys[5], **pred_dict)
I’ve tried all available autoguides; step_size & init_scale from 0.0001 to 1.0; with no major difference.
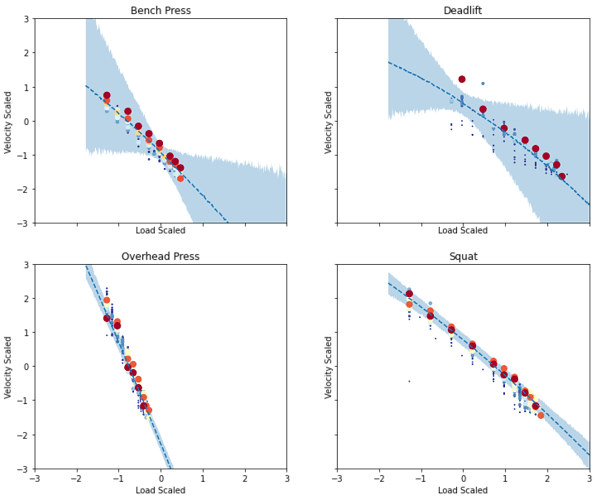
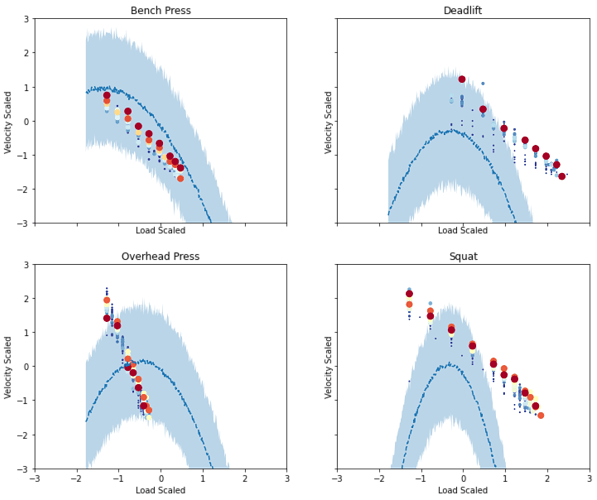
 . num_steps 1e5 works, but no luck in combo with step_size 1e-5.
. num_steps 1e5 works, but no luck in combo with step_size 1e-5.