Thanks for the useful replies; I am just writing to follow-up on this question since I’m still having troubles implementing this in an effective way in Pyro. In summary, I’m able to write an explicit model (looping over all variables) but struggling to write the equivalent model using the plate contexts.
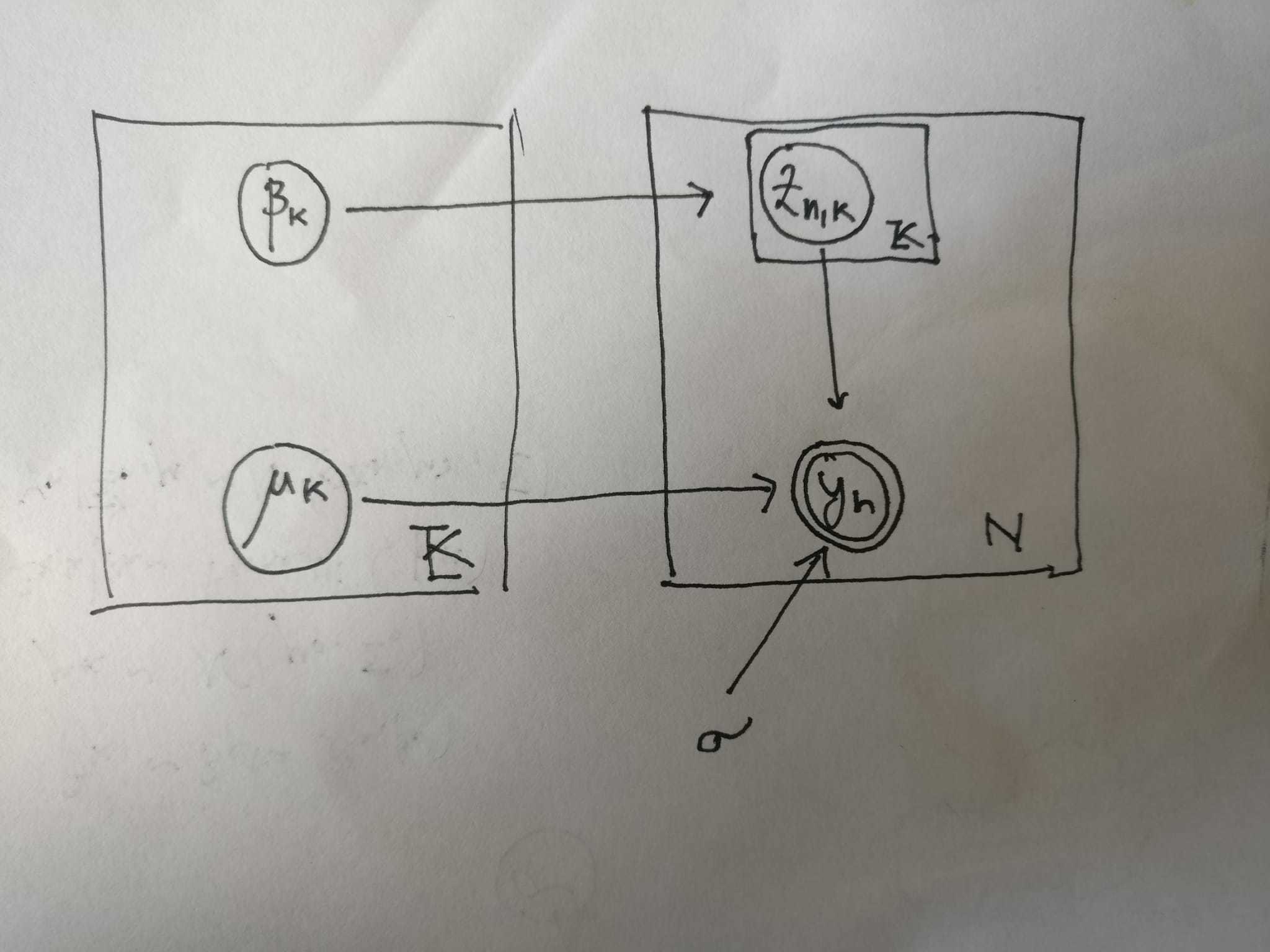
To be concrete, the plate model I have in mind is the following:
I am facing the following issue: I write down the non-plate, sequential version of the model, I get something that seems to be compiling, however, the corresponding “plate notation” seems to be having issues. Any suggestion to make the code correct (I eventually would like to move to the case in which the data is in dimension d, i.e. i want to be using torch.matmul(assignments, mu) in place of torch.dot(assignment, mu)) would be greatly appreciated!
For completeness I attach the actual code I am running. For the “sequential”, non-plate version:
num_components = 2 # Fixed number of components.
def model(data):
N = len(data)
scale = torch.tensor(1.0)
# after all plates, weights and locs will be tensors of size (num_components,)
weights = torch.zeros(num_components)
locs = torch.zeros(num_components)
for k in range(num_components):
weights[k] = pyro.sample('weights_{}'.format(k), dist.Beta(0.5, 0.5))
locs[k] = pyro.sample('locs_{}'.format(k), dist.Normal(0., 10.))
for n in range(N):
Z = torch.zeros(num_components)
for k in range(num_components):
Z[k] = pyro.sample('assignments_{}_{}'.format(n,k), dist.Bernoulli(weights[k]))
pyro.sample('obs_{}'.format(n), dist.Normal(torch.dot(Z, locs), scale),obs=data[n])
def guide(data):
N = len(data)
# register variational parameters, with the initial values
kappa = pyro.param(‘kappa’, lambda: dist.Uniform(0, 2).sample([num_components]), constraint=constraints.positive)
tau = pyro.param(‘tau’, lambda: dist.Normal(torch.zeros(1), 1).sample([num_components]))
phi = pyro.param(‘phi’, lambda: dist.Beta(1.,1.).sample([N, num_components]), constraint=constraints.unit_interval)
# register variational distributions
for k in range(num_components):
pyro.sample('weights_{}'.format(k),dist.Beta(1.0,kappa[k]))
pyro.sample('locs_{}'.format(k), dist.Normal(tau[k], 1.0))
for n in range(N):
for k in range(num_components):
pyro.sample('assignments_{}_{}'.format(n,k), dist.Bernoulli(phi[n,k]))
optim = pyro.optim.Adam({‘lr’: 0.1, ‘betas’: [0.8, 0.99]})
elbo = Trace_ELBO()
svi = SVI(model, guide, optim, loss=elbo)
n_steps = 100
for step in range(n_steps):
svi.step(data)
The parallel, plate version:
def plate_model(data):
N = len(data)
scale = torch.tensor(1.0)
with pyro.plate(“components”, num_components):
weights = pyro.sample(‘weights’, dist.Beta(0.5, 0.5))
locs = pyro.sample(‘locs’, dist.Normal(0., 10.))
with pyro.plate('data', len(data), dim = -2):
# Local variables.
Z = pyro.sample('assignments', dist.Bernoulli(weights))
pyro.sample('obs', dist.Normal(torch.dot(Z, locs), scale), obs=data)
def plate_guide(data):
# register variational parameters, with the initial values
kappa = pyro.param('kappa', lambda: dist.Uniform(0, 2).sample([num_components]), constraint=constraints.positive)
tau = pyro.param('tau', lambda: dist.Normal(torch.zeros(1), 1).sample([num_components]))
phi = pyro.param('phi', lambda: dist.Beta(1.,1.).sample([N, num_components]), constraint=constraints.unit_interval)
with pyro.plate("components", num_components):
weights = pyro.sample('weights', dist.Beta(0.5, kappa))
locs = pyro.sample('locs', dist.Normal(tau, 10.))
with pyro.plate('data', len(data), dim = -2):
Z = pyro.sample('assignments', dist.Bernoulli(phi))
optim = pyro.optim.Adam({‘lr’: 0.1, ‘betas’: [0.8, 0.99]})
elbo = Trace_ELBO()
svi = SVI(plate_model, plate_guide, optim, loss=elbo)
for i in tqdm(range(200)):
loss = svi.step(data)
I get the following error:
RuntimeError: 1D tensors expected, got 2D, 2D tensors at /opt/conda/conda-bld/pytorch_1595629395347/work/aten/src/TH/generic/THTensorEvenMoreMath.cpp:83
Trace Shapes:
Param Sites:
Sample Sites:
components dist |
value 2 |
weights dist 2 |
value 2 |
locs dist 2 |
value 2 2 |
data dist |
value 100 |
assignments dist 100 2 |
value 100 2 |
If I remove the dim = -2 in the plate notation I get a different error (here I ran on data with data.shape[0] = 100):
ValueError: Shape mismatch inside plate(‘data’) at site assignments dim -1, 100 vs 2
Trace Shapes:
Param Sites:
kappa 2
tau 2 1
phi 100 2
Sample Sites:
components dist |
value 2 |
weights dist 2 |
value 2 |
locs dist 2 2 |
value 2 2 |
data dist |
value 100 |
Another issue I’ve had is related to initialization in self-designed guides: I tried to look for examples of guides with initializations (in the spirit of init_loc_fn() in Gaussian Mixture Model — Pyro Tutorials 1.8.6 documentation) but I couldn’t find any.
Thanks!