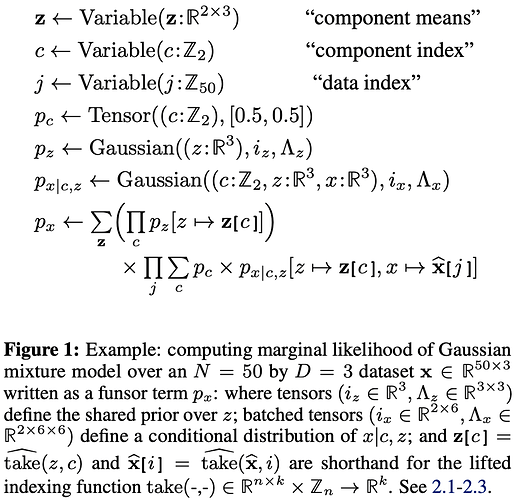
Thanks, that helps. The stumbling block was expecting some kind of differentiation between z and x.
Now I still have remaining questions about c in that funsor. How does the system determine that the Gaussian is over z,x but not c? Does it do it based on the fact that c is integer-typed? Could c have been real-typed but not be considered one of the Gaussian-distributed variables?
Also like I mentioned above, I am unclear about the need to have c in p_{x|c,z}. Here is how I read the example:
\sum_c p_c \times p_{x|c,z}[z \mapsto {\bf z}[c], x \mapsto \widehat{\bf x}[j]] is equivalent to
p_{c} [c \mapsto 0] \times p_{x|c,z}[z \mapsto {\bf z}[0], x \mapsto \widehat{\bf x}[j]] + p_{c} [c \mapsto 1] \times p_{x|c,z}[z \mapsto {\bf z}[1], x \mapsto \widehat{\bf x}[j]]
From what I understand, p_{x|c,z}[z \mapsto {\bf z}[0], x \mapsto \widehat{\bf x}[j]] evaluates to a funsor with a single {\mathbb Z}^2 dimension c containing two Gaussians, both of which defined over {\bf z}[0]. This confuses me because I would expect either to have a single Gaussian, or to have two Gaussians, each of which corresponding to one of the Gaussian components, not to have both defined on {\bf z}[0]. We also have the analogous situation in the second term for {\bf z}[1].
If I were to write the model myself with my current understanding, I would have written something like:
\sum_c p_c[c \mapsto c] \times p_{x|z}[z \mapsto {\bf z}[c], x \mapsto \widehat{\bf x}[j]],
with a previously defined
p_{x|z} \leftarrow \operatorname{Gaussian}((z:{\mathbb R}^3, x: {\mathbb R}^3), i_x, \Lambda_x).
(The p_c[c \mapsto c] is my attempt of reducing {\mathbb Z}^2-shaped p_c to a single probability, but I am not sure that works, or if p_c[c] would make more sense instead, or neither).
Alternatively, I would try writing a vectorized form as in:
p_c \times p_{x|z}[z \mapsto {\bf z}, x \mapsto \widehat{\bf x}[j]].
Can you identify what I am missing?