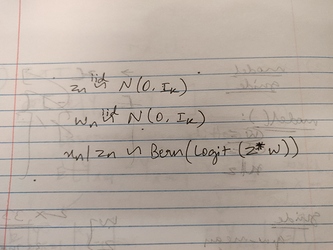I’m working on Probabilistic PCA where the model assumes the following data generating process:
This model in Pyro:
def ppca_model(data):
data_dim = data.shape[1]
latent_dim = 2
num_datapoints = data.shape[0]
w_mean0 = torch.zeros([latent_dim, data_dim])
w_std0 = torch.ones([latent_dim, data_dim])
z_mean0 = torch.zeros([num_datapoints, latent_dim])
z_std0 = torch.ones([num_datapoints, latent_dim])
w = pyro.sample("w", pyro.distributions.Normal(loc = w_mean0,
scale = w_std0))
z = pyro.sample("z", pyro.distributions.Normal(loc = z_mean0,
scale = z_std0))
linear_exp = torch.exp(torch.matmul(z, w))
x = pyro.sample("x", pyro.distributions.Bernoulli(probs = linear_exp/(1+linear_exp)))
Given a one-hot-encoded data of dimension 3000 x 500, I try to infer latent variable z of dimension 3000 x 2.
Inference is attempted by HMC:
from pyro.infer.mcmc import HMC, MCMC
hmc_kernel = HMC(ppca_model, step_size=0.0855, num_steps=4)
mcmc_run = MCMC(hmc_kernel, num_samples=500, warmup_steps=100).run(x_new)
This breaks with the error message:
RuntimeError Traceback (most recent call last)
in
8 ppca_model(x_new)
9
—> 10 mcmc_run = MCMC(hmc_kernel, num_samples=500, warmup_steps=100).run(x_new)~\Anaconda3\lib\site-packages\pyro\infer\abstract_infer.py in run(self, *args, **kwargs)
221 self._reset()
222 with poutine.block():
→ 223 for i, vals in enumerate(self._traces(*args, **kwargs)):
224 if len(vals) == 2:
225 chain_id = 0~\Anaconda3\lib\site-packages\pyro\infer\mcmc\mcmc.py in _traces(self, *args, **kwargs)
266
267 def _traces(self, *args, **kwargs):
→ 268 for sample in self.sampler._traces(*args, **kwargs):
269 yield sample
270~\Anaconda3\lib\site-packages\pyro\infer\mcmc\mcmc.py in _traces(self, *args, **kwargs)
204 progress_bar = ProgressBar(self.warmup_steps, self.num_samples, disable=self.disable_progbar)
205 self.logger = initialize_logger(self.logger, logger_id, progress_bar, log_queue)
→ 206 self.kernel.setup(self.warmup_steps, *args, **kwargs)
207 trace = self.kernel.initial_trace
208 with optional(progress_bar, not is_multiprocessing):~\Anaconda3\lib\site-packages\pyro\infer\mcmc\hmc.py in setup(self, warmup_steps, *args, **kwargs)
380 self._args = args
381 self._kwargs = kwargs
→ 382 self._initialize_model_properties()
383
384 def cleanup(self):~\Anaconda3\lib\site-packages\pyro\infer\mcmc\hmc.py in _initialize_model_properties(self)
353 self._trace_prob_evaluator = TraceEinsumEvaluator(trace,
354 self._has_enumerable_sites,
→ 355 self.max_plate_nesting)
356 if site_value is not None:
357 mass_matrix_size = sum(self._r_numels.values())~\Anaconda3\lib\site-packages\pyro\infer\mcmc\util.py in init(self, model_trace, has_enumerable_sites, max_plate_nesting)
158 self._enum_dims = set()
159 self.ordering = {}
→ 160 self._populate_cache(model_trace)
161
162 def _populate_cache(self, model_trace):~\Anaconda3\lib\site-packages\pyro\infer\mcmc\util.py in _populate_cache(self, model_trace)
170 raise ValueError("Finite value required formax_plate_nestingwhen model "
171 “has discrete (enumerable) sites.”)
→ 172 model_trace.compute_log_prob()
173 model_trace.pack_tensors()
174 for name, site in model_trace.nodes.items():~\Anaconda3\lib\site-packages\pyro\poutine\trace_struct.py in compute_log_prob(self, site_filter)
214 if “log_prob” not in site:
215 try:
→ 216 log_p = site[“fn”].log_prob(site[“value”], *site[“args”], **site[“kwargs”])
217 except ValueError:
218 _, exc_value, traceback = sys.exc_info()~\Anaconda3\lib\site-packages\torch\distributions\bernoulli.py in log_prob(self, value)
92 if self._validate_args:
93 self._validate_sample(value)
—> 94 logits, value = broadcast_all(self.logits, value)
95 return -binary_cross_entropy_with_logits(logits, value, reduction=‘none’)
96~\Anaconda3\lib\site-packages\torch\distributions\utils.py in broadcast_all(*values)
35 break
36 values = [v if torch.is_tensor(v) else new_tensor(v) for v in values]
—> 37 return torch.broadcast_tensors(*values)
38
39~\Anaconda3\lib\site-packages\torch\functional.py in broadcast_tensors(*tensors)
60 [0, 1, 2]])
61 “”"
—> 62 return torch._C._VariableFunctions.broadcast_tensors(tensors)
63
64RuntimeError: The size of tensor a (500) must match the size of tensor b (2) at non-singleton dimension 1
At some point, it tries to broadcast tensors of dimension 3000 x 500 and 2. How do I solve this error? Or can somebody recommend any other inference technique that can work on this model?