I realize that it may be hard to do much troubleshooting without the full model code, but here’s the seemingly relevant info I can easily give:
I have a hyperparameter mu_tau for mu defined roughly like this:
with reparam(config={"mu_tau": TransformReparam()}):
mu_tau = (
ny.sample(
f"mu_tau",
dist.TransformedDistribution(dist.HalfNormal(scale=1), AffineTransform(loc=0, scale=0.1)),
)
)
with reparam(config={"mu": TransformReparam()}):
mu = ny.sample(
"mu",
dist.TransformedDistribution(
dist.Normal(loc=0, scale=1).expand((shape.n_endog,)),
LowerCholeskyAffine(loc=jnp.zeros(shape.n_endog), scale_tril=mu_chol * mu_tau),
),
)
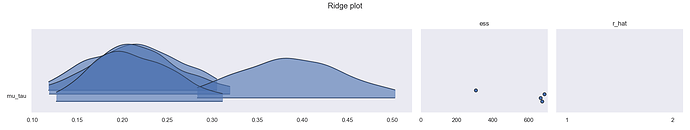
This model works fine (0 divergences, r hats at 1) around 70% of the time. But for some PRNGKeys passed to mcmc.run, one chain is an outlier from the rest (also happens to ~2 chains when I bump up to 8 chains). Here are the diagnostic graphs.
Latent posterior distributions for each chain:
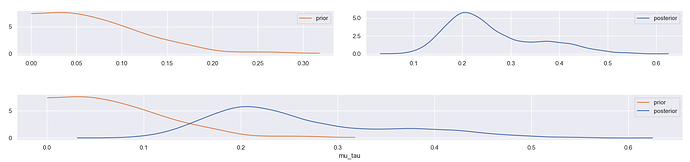
Prior and posterior distributions:
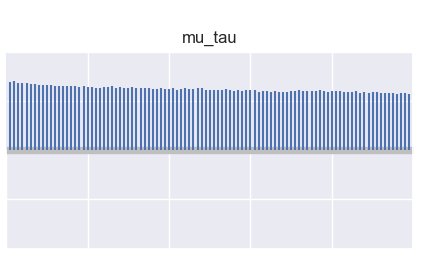
Chain autocorrelation:
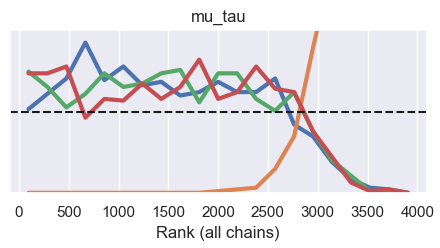
Rank plot:
Trace plot:
Does anyone have any guesses as to what’s going on or what additional diagnostic information I should seek?