Hello,
TL;DR: I have a model that work with PyMC3 that I want to refactor to Pyro but the Pyro implementation is quite off compared to the PyMC3 one.
Background
I have been working on a model based on spline regression that I have implemented in PyMC3 and I would like to re-implement it in Pyro for scalability and also to evolve it. Basically I have different classes c (that I know) and for each classes I have a sequence of (x, y) that I am modelling using a spline regression as follow:
import pymc3 as pm
import scipy as sp
from theano import shared
# dummy data:
n = 100
def foo(x):
return np.sin(x * np.pi) + np.sin(x * np.pi * 2)
x = np.random.uniform(size = n)
e = np.random.normal(0, .1, n)
y = foo(x) + e
# spline regressors:
N_KNOT = 10
knots = np.linspace(0, 1, N_KNOT)
basis_funcs = sp.interpolate.BSpline(knots, np.eye(N_KNOT), k=1) # first order splines
trend_x = basis_funcs(x)
# PyMC3 model for a single class
trend_x_ = shared(trend_x)
n_ts = trend_x.shape[1]
with pm.Model() as model_spline:
σ_a = pm.HalfCauchy('σ_a', 5.)
a0 = pm.Normal('a0', 0., 10.)
Δ_a = pm.Normal('Δ_a', 0., 1., shape=n_ts)
a = pm.Deterministic('a', a0 + (σ_a * Δ_a).cumsum())
mu = trend_x_.dot(a)
σ = pm.HalfCauchy('σ', 5.)
obs = pm.Normal('obs', mu = mu, sd = σ, observed=y)
with model_spline:
inference_spline = pm.ADVI()
approx_spline = pm.fit(n=25000, method=inference_spline)
trace_spline = approx_spline.sample(draws=1000)
And for the dummy data, the results are quite fitting:
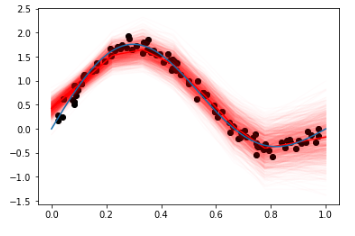
Pyro implementation
To simplify the management of the different classes, I wanted to leverage the Embedding layer and I replicated the implementation of the spline regression in PyMC3 in Pytorch:
import torch
import torch.nn as nn
from torch.autograd import Variable
class SplineRegression(nn.Module):
def __init__(self, n_nodes, n_classes):
super(SplineRegression, self).__init__()
self.n_nodes = n_nodes
self.n_classes = n_classes
self.σ_a = nn.Embedding(self.n_classes, 1)
self.Δ_a = nn.Embedding(self.n_classes, self.n_nodes)
self.a0 = nn.Embedding(self.n_classes, 1)
self.σ = nn.Embedding(self.n_classes, 1)
def forward(self, c, t):
a0 = self.a0(c) # <- retrieve the value for the corresponding class/category
Δ_a = self.Δ_a(c) # <- retrieve the values for the corresponding class/category
σ_a = self.σ_a(c) # <- retrieve the value for the corresponding class/category
σ = self.σ(c) # <- retrieve the value for the corresponding class/category
a = torch.cumsum(Δ_a, 1).mul(σ_a.reshape(-1, 1)) + a0
return torch.sum(t * a, 1).reshape(-1, 1), σ.reshape(-1, 1)
And I then created a Bayesian implementation with “lifted” modules, trying to use the same distributions as in the PyMC3 implementation:
import pyro
import pyro.distributions as dist
from pyro.infer import SVI, Trace_ELBO
from pyro.optim import Adam
class BayesianSplineRegression(nn.Module):
def __init__(self, n_nodes, n_classes):
super(BayesianSplineRegression, self).__init__()
self.regression = SplineRegression(n_nodes, n_classes)
self.n_nodes = n_nodes
self.n_classes = n_classes
pyro.clear_param_store()
def model(self, c, t, y):
σ_a_prior = dist.HalfCauchy(Variable(5. * torch.ones(self.n_classes, 1)))
σ_prior = dist.HalfCauchy(Variable(5. * torch.ones(self.n_classes, 1)))
Δ_a_prior = dist.Normal(
Variable(torch.zeros(self.n_classes, self.n_nodes)),
Variable(torch.ones(self.n_classes, self.n_nodes))
)
a0_prior = dist.Normal(
Variable(torch.zeros(self.n_classes, 1)),
Variable(10.*torch.ones(self.n_classes, 1))
)
priors = {
'σ_a.weight': σ_a_prior.to_event(2),
'Δ_a.weight': Δ_a_prior.to_event(2),
'a0.weight': a0_prior.to_event(2),
'σ.weight': σ_prior.to_event(2),
}
lifted_module = pyro.random_module("regression", self.regression, priors)
lifted_regression = lifted_module()
with pyro.plate("map", len(c)):
mu, sig = lifted_regression(c, t)
pyro.sample("obs", dist.Normal(mu, sig).to_event(2), obs=y)
return mu, sig
def guide(self, c=None, t=None, y=None):
a0_loc = pyro.param('a0_loc', torch.zeros(self.n_classes, 1))
a0_scale = pyro.param('a0_scale', torch.ones(self.n_classes, 1), constraint=constraints.positive)
σ_a_loc = pyro.param('σ_a_loc', torch.ones(self.n_classes, 1), constraint=constraints.positive)
σ_loc = pyro.param('σ_loc', torch.ones(self.n_classes, 1), constraint=constraints.positive)
Δ_a_loc = pyro.param('Δ_a_loc', torch.zeros(self.n_classes, self.n_nodes))
Δ_a_scale = pyro.param('Δ_a_scale', torch.ones(self.n_classes, self.n_nodes), constraint=constraints.positive)
σ_a = dist.HalfCauchy(σ_a_loc)
σ = dist.HalfCauchy(σ_loc)
a0 = dist.Normal(a0_loc, a0_scale)
Δ_a = dist.Normal(Δ_a_loc, Δ_a_scale)
dists = {
'σ_a.weight': σ_a.to_event(2),
'Δ_a.weight': Δ_a.to_event(2),
'a0.weight': a0.to_event(2),
'σ.weight': σ.to_event(2),
}
lifted_module = pyro.random_module("regression", self.regression, dists)
return lifted_module()
def fit(self, c, t, y, lr=0.001, n_iter=10):
pyro.clear_param_store()
optim = Adam({"lr": lr})
elbo = Trace_ELBO()
self.svi = SVI(self.model, self.guide, optim, loss=elbo)
losses = []
for i in range(n_iter):
losses.append(self.svi.step(c, t, y)/len(c))
return losses
then, fit:
c_tch = torch.LongTensor([0]*n)
t_tch = torch.Tensor(trend_x)
y_tch = torch.Tensor(y)
model = BayesianSplineRegression(n_ts, 1)
losses = model.fit(c_tch, t_tch, y_tch, lr=0.1, n_iter=25000)
I then try to look at the fitted results by sampling models:
x_plot = np.linspace(0, 1, 299)
t_tch_pred = torch.FloatTensor(basis_funcs(x_plot))
c_tch_pred = torch.LongTensor([0]*x_plot.shape[0])
M = np.zeros((1000, len(x_plot)))
for i in range(1000):
m_, _ = model.guide()(c_tch_pred, t_tch_pred)
m_ = m_.detach().cpu().numpy().ravel()
M[i,:] = m_
plt.plot(x_plot, m_, alpha=.01, c='red')
plt.plot(x_plot, M.mean(0), c='red')
plt.plot(x_plot, foo(x_plot))
plt.scatter(x, y, c='black');
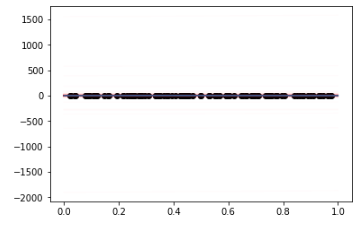
And the results is quite different from my other implementation. I am wondering whether I have a problem with the implementation of the guide function and I have tried to use the auto-guide function like AutoDiagonalNormal but I also get odd results. Any ideas on what I might have done wrong?
Thanks in advance!